
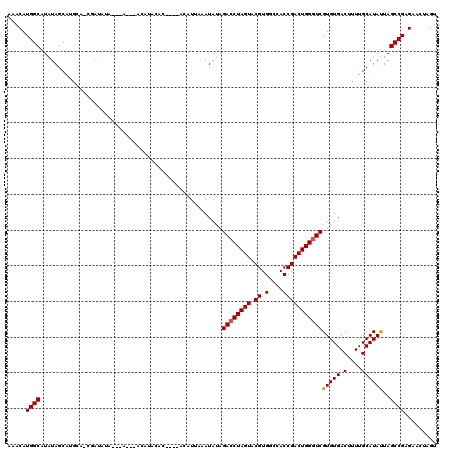
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,259,323 – 3,259,463 |
| Length | 140 |
| Max. P | 0.999933 |

| Location | 3,259,323 – 3,259,423 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3259323 100 + 22407834 AAACAUGGCAUAUAGCAUGCA------UACACAC--------------ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGU .....((((...(((.(((((------....(((--------------((((.....)).((((((((.((.(....)))))))))))))))).....))))).)))))))......... ( -28.30) >DroSec_CAF1 85899 106 + 1 AAACAUGGCAUAUAGCAUGCAGCGAUAUAUGUAU--------------ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGU .....((((.....((.....))...(((((((.--------------.......((((.((((((((.((.(....)))))))))))))))......)))))))..))))......... ( -26.64) >DroSim_CAF1 89021 119 + 1 AAACAUGGCAUAUAGCAUGCAGCGAUAUAUGUAUGUACAUACACACACACAUUAAAUAUAGACCUAGUACGUGACCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACU-GU .....((((.....(((((((........))))))).........(((((((.....)).((((((((.((.......)))))))))))))))..............))))......-.. ( -31.00) >DroEre_CAF1 82740 107 + 1 AAACAUGGCAUAUAGCUGGUG-GGAUAU--------ACAUACAC----ACAUAUUAUAUAGACCUAGUACGUGUCCACCGACUGGGUCGUGUGACUUUUGCAUACUAGCCGGGAACUAGU .....((((((((((...(((-......--------.......)----))...)))))).((((((((.((.(....)))))))))))(((((.(....))))))..))))......... ( -28.02) >DroYak_CAF1 82111 108 + 1 AAACAUGGCAUGUAGCAGGAAUGUAUGU--------ACAUACAC----ACAUAUUAUAUAGACCUAGUACGUGUCCACCGACUGGAUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGU .....((((..(((((((((((((.(((--------....))).----)))).((((((.((.(((((.((.(....)))))))).)))))))).)))))).)))..))))......... ( -24.10) >consensus AAACAUGGCAUAUAGCAUGCA_CGAUAUA___A___ACAUACAC____ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGU .....((((...................................................((((((((.((.(....)))))))))))(((((.(....))))))..))))......... (-21.18 = -21.22 + 0.04)


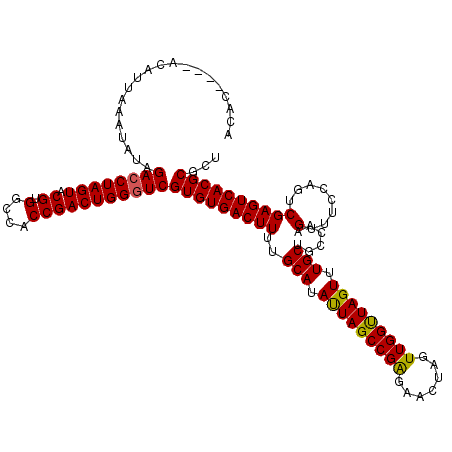
| Location | 3,259,351 – 3,259,463 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -37.22 |
| Energy contribution | -36.94 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3259351 112 + 22407834 --------ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGUUGGUUAGUUUGCUGCCAAGUUUCCAGUCGAGUCACGCGCU --------............((((((((.((.(....)))))))))))(((((((((..(((.(((((((((.......))))))))).)))......(........))))))))))... ( -37.70) >DroSec_CAF1 85933 112 + 1 --------ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGUUGGUUAGUUUGCUGCCAAGUUUCCAGUCGAGUCACGCGCU --------............((((((((.((.(....)))))))))))(((((((((..(((.(((((((((.......))))))))).)))......(........))))))))))... ( -37.70) >DroSim_CAF1 89061 119 + 1 ACACACACACAUUAAAUAUAGACCUAGUACGUGACCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACU-GUUGGUUAGUUUGCUGCCAAGUUUCCAGUCGAGUCACGCGCU .........................((..((((((...((((((((......(((((..(((.(((((((((.....-.))))))))).)))....))))))))))))).))))))..)) ( -37.60) >DroEre_CAF1 82771 116 + 1 ACAC----ACAUAUUAUAUAGACCUAGUACGUGUCCACCGACUGGGUCGUGUGACUUUUGCAUACUAGCCGGGAACUAGUUGGCUAGUUUGCUGCCAAGUUUCUAGUCGAGUCACGCGCU ....----............((((((((.((.(....)))))))))))(((((((((..(((.(((((((((.......))))))))).)))......(........))))))))))... ( -41.20) >DroYak_CAF1 82143 116 + 1 ACAC----ACAUAUUAUAUAGACCUAGUACGUGUCCACCGACUGGAUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGUUGGUUAGUUUGCUGCCAAGUUUCCAGUCGAGUCACGCGCU ....----.................((..((((.(...((((((((......(((((..(((.(((((((((.......))))))))).)))....))))))))))))).).))))..)) ( -34.10) >consensus ACAC____ACAUUAAAUAUAGACCUAGUACGUGGCCACCGACUGGGUCGUGUGACUUUUGCAUAUUAGCCGAGAACUAGUUGGUUAGUUUGCUGCCAAGUUUCCAGUCGAGUCACGCGCU ....................((((((((.((.(....)))))))))))(((((((((..(((.(((((((((.......))))))))).)))......(........))))))))))... (-37.22 = -36.94 + -0.28)

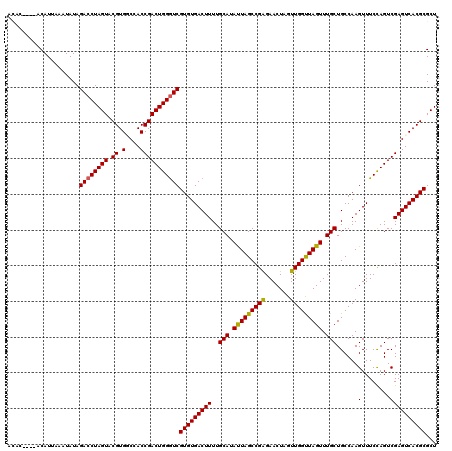
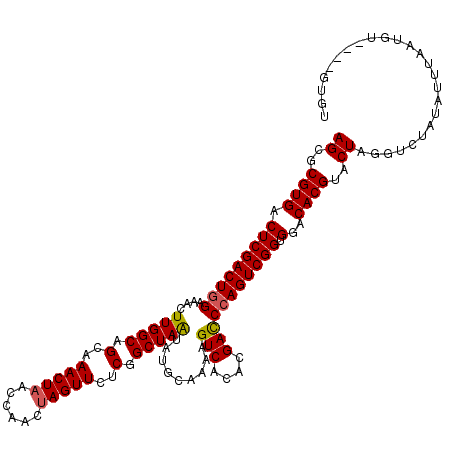
| Location | 3,259,351 – 3,259,463 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3259351 112 - 22407834 AGCGCGUGACUCGACUGGAAACUUGGCAGCAAACUAACCAACUAGUUCUCGGCUAAUAUGCAAAAGUCACACGACCCAGUCGGUGGCCACGUACUAGGUCUAUAUUUAAUGU-------- ((..((((.(((((((((....(((((.(..(((((......)))))..).))))).........(((....))))))))))).)..))))..)).................-------- ( -30.20) >DroSec_CAF1 85933 112 - 1 AGCGCGUGACUCGACUGGAAACUUGGCAGCAAACUAACCAACUAGUUCUCGGCUAAUAUGCAAAAGUCACACGACCCAGUCGGUGGCCACGUACUAGGUCUAUAUUUAAUGU-------- ((..((((.(((((((((....(((((.(..(((((......)))))..).))))).........(((....))))))))))).)..))))..)).................-------- ( -30.20) >DroSim_CAF1 89061 119 - 1 AGCGCGUGACUCGACUGGAAACUUGGCAGCAAACUAACCAAC-AGUUCUCGGCUAAUAUGCAAAAGUCACACGACCCAGUCGGUGGUCACGUACUAGGUCUAUAUUUAAUGUGUGUGUGU ((..((((((((((((((....(((((.(..((((.......-))))..).))))).........(((....))))))))))..)))))))..)).....(((((.....)))))..... ( -32.60) >DroEre_CAF1 82771 116 - 1 AGCGCGUGACUCGACUAGAAACUUGGCAGCAAACUAGCCAACUAGUUCCCGGCUAGUAUGCAAAAGUCACACGACCCAGUCGGUGGACACGUACUAGGUCUAUAUAAUAUGU----GUGU .((((((((((.(.((((....))))).(((.(((((((...........))))))).)))...))))))..((((.(((((.......)).))).))))..........))----)).. ( -37.60) >DroYak_CAF1 82143 116 - 1 AGCGCGUGACUCGACUGGAAACUUGGCAGCAAACUAACCAACUAGUUCUCGGCUAAUAUGCAAAAGUCACACGAUCCAGUCGGUGGACACGUACUAGGUCUAUAUAAUAUGU----GUGU .(((((((...((((((((....((((.(..(((((......)))))..).((......))....)))).....))))))))((((((.(......)))))))....)))))----)).. ( -33.80) >consensus AGCGCGUGACUCGACUGGAAACUUGGCAGCAAACUAACCAACUAGUUCUCGGCUAAUAUGCAAAAGUCACACGACCCAGUCGGUGGACACGUACUAGGUCUAUAUUUAAUGU____GUGU ((..((((.(((((((((....(((((.(..(((((......)))))..).))))).........(((....))))))))))).)..))))..))......................... (-27.52 = -27.60 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:57 2006