
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,247,767 – 3,247,925 |
| Length | 158 |
| Max. P | 0.994454 |

| Location | 3,247,767 – 3,247,887 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -25.80 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3247767 120 + 22407834 CUAGCACUGACAGUUACAAUUGCUGGUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAG ...(((.(((((((.......))).))))...(((((((......((((((((((..(((.(((....))).))).)))))))))).........))))))).))).............. ( -30.26) >DroSec_CAF1 81815 120 + 1 CUAGCAUGGACAGAUACAAUUGCUGGUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGAAGAAUCAAAAG ((((((((........))..)))))).....((((.(((((((..((((((((((..(((.(((....))).))).))))))))))...........)))))))...))))......... ( -31.82) >DroSim_CAF1 84947 120 + 1 CUAGCAUGGACAGAUACAAUUGCUGGUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAG ............(((....((((((..(((..(((((((......((((((((((..(((.(((....))).))).)))))))))).........)))))))))))))))).)))..... ( -29.66) >DroEre_CAF1 78584 120 + 1 CUAGCACUGACAGAUACAAUUGCUGCUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCGAAAUGGGCAGCCAGAAACCGCAACAAAGUUCUAUGGAAAUGCAGCAGAAUCAAAAG ............(((....((((((((((((....(((((.....((((((((((..(((.((......)).))).))))))))))...))))).)))))....))))))).)))..... ( -27.10) >DroYak_CAF1 77841 120 + 1 CUAGCACUGACAGAUACAAUUGCUGGUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCCCAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAG ............(((....((((((..(((..(((((((......((((((((((..(((.(((....))).))).)))))))))).........)))))))))))))))).)))..... ( -28.66) >consensus CUAGCACUGACAGAUACAAUUGCUGGUCAUACUUCAAUUUCAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAG ...................((((((..(((..(((((((......((((((((((..(((.(((....))).))).)))))))))).........))))))))))))))))......... (-25.80 = -26.44 + 0.64)

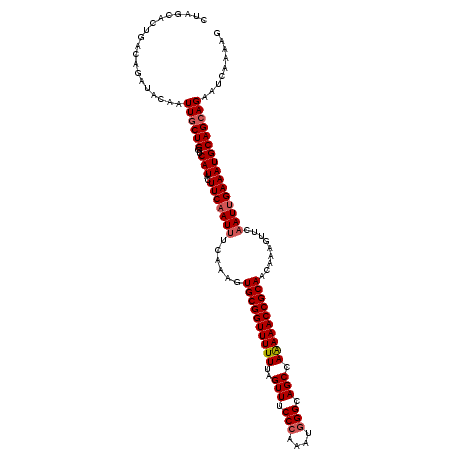

| Location | 3,247,767 – 3,247,887 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.74 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3247767 120 - 22407834 CUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGACCAGCAAUUGUAACUGUCAGUGCUAG ...........((((((.((((((((.........(((((((((((...(((....)))...)).)))))))))(((((......)))))........)))))).)).)).))))..... ( -30.50) >DroSec_CAF1 81815 120 - 1 CUUUUGAUUCUUCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGACCAGCAAUUGUAUCUGUCCAUGCUAG ...................((((((.......((.(((((((((((...(((....)))...)).))))))))))).....))))))((((((..(((.........)))..)))))).. ( -29.50) >DroSim_CAF1 84947 120 - 1 CUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGACCAGCAAUUGUAUCUGUCCAUGCUAG ..........(((((.(((((((((.......((.(((((((((((...(((....)))...)).))))))))))).....))))))))).....)))))...((((......))))... ( -30.20) >DroEre_CAF1 78584 120 - 1 CUUUUGAUUCUGCUGCAUUUCCAUAGAACUUUGUUGCGGUUUCUGGCUGCCCAUUUCGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGAGCAGCAAUUGUAUCUGUCAGUGCUAG ..............(((((...(((((((.((((((((((((((((.........))))))))).......((.(((((......))))).)).)))))))..)).))))).)))))... ( -28.40) >DroYak_CAF1 77841 120 - 1 CUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUGGGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGACCAGCAAUUGUAUCUGUCAGUGCUAG ..........(((((.(((((((((.......((.(((((((((((...(((....)))...)).))))))))))).....))))))))).....)))))...((((......))))... ( -30.20) >consensus CUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUGAAAUUGAAGUAUGACCAGCAAUUGUAUCUGUCAGUGCUAG ..........(((((.(((((((((.......((.(((((((((((...(((....)))...)).))))))))))).....))))))))).....)))))...((((......))))... (-25.70 = -26.74 + 1.04)

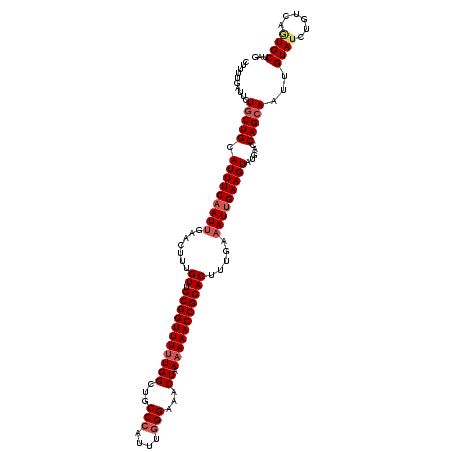
| Location | 3,247,807 – 3,247,925 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3247807 118 + 22407834 CAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAGAAAAAUCACCCAAAUACC--AGUGGCAUAGAGAAAAAUGC .....((((((((((..(((.(((....))).))).))))))))))................((((.......((....)).....(((.........--.)))))))............ ( -22.30) >DroSec_CAF1 81855 118 + 1 CAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGAAGAAUCAAAAGAAAAAUCACCCAAAUACC--AGUGGCAUAGAGAAAAAUGC .....((((((((((..(((.(((....))).))).))))))))))................((((.((....))...........(((.........--.)))))))............ ( -22.40) >DroSim_CAF1 84987 117 + 1 CAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAGAAAAAUCACCCAAAUAAC--AGUGGCAUAGAGAA-AAUGC .....((((((((((..(((.(((....))).))).))))))))))................((((.......((....)).....(((.........--.)))))))......-..... ( -22.30) >DroEre_CAF1 78624 117 + 1 CAAAGUGCGGUUUUUUAGUUUCCGAAAUGGGCAGCCAGAAACCGCAACAAAGUUCUAUGGAAAUGCAGCAGAAUCAAAAGAAAAAUCGUCCAAAUACC--AGUGGCAUAGAAA-AAAUGC .....((((((((((..(((.((......)).))).))))))))))......(((((((....................((....))..(((......--..)))))))))).-...... ( -22.70) >DroYak_CAF1 77881 119 + 1 CAAAGUGCGGUUUUUUAGUUUCCCCAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAGAAAAAUCACACAAAUACCCAAAUGGCAUAGAAA-AAAUGC .....((((((((((..(((.(((....))).))).)))))))))).....((((..(((.....)))..))))..............................((((.....-..)))) ( -20.40) >consensus CAAAGUGCGGUUUUUUAGUUUCCCAAAUGGGCAGCCAAAAACCGCAACAAAGUUCAAUUGAAAUGCAGCAGAAUCAAAAGAAAAAUCACCCAAAUACC__AGUGGCAUAGAGAAAAAUGC .....((((((((((..(((.(((....))).))).)))))))))).....((((..(((.....)))..))))..............................((((........)))) (-19.64 = -19.88 + 0.24)

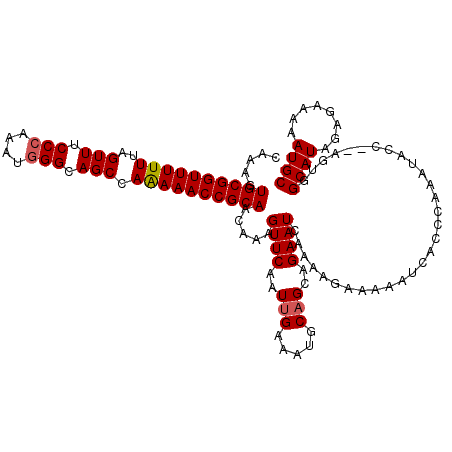
| Location | 3,247,807 – 3,247,925 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3247807 118 - 22407834 GCAUUUUUCUCUAUGCCACU--GGUAUUUGGGUGAUUUUUCUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUG ((((........))))((((--.(....).))))....(((..((((............))))..)))......((((((((((((...(((....)))...)).))))))))))..... ( -27.90) >DroSec_CAF1 81855 118 - 1 GCAUUUUUCUCUAUGCCACU--GGUAUUUGGGUGAUUUUUCUUUUGAUUCUUCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUG ((((........))))((((--.(....).))))....(((..((((............))))..)))......((((((((((((...(((....)))...)).))))))))))..... ( -27.90) >DroSim_CAF1 84987 117 - 1 GCAUU-UUCUCUAUGCCACU--GUUAUUUGGGUGAUUUUUCUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUG ((((.-......))))((((--........))))....(((..((((............))))..)))......((((((((((((...(((....)))...)).))))))))))..... ( -26.40) >DroEre_CAF1 78624 117 - 1 GCAUUU-UUUCUAUGCCACU--GGUAUUUGGACGAUUUUUCUUUUGAUUCUGCUGCAUUUCCAUAGAACUUUGUUGCGGUUUCUGGCUGCCCAUUUCGGAAACUAAAAAACCGCACUUUG (((...-.(((((((.....--.(((...((((((........))).)))...))).....)))))))...)))((((((((.(((....)))....(....)....))))))))..... ( -22.30) >DroYak_CAF1 77881 119 - 1 GCAUUU-UUUCUAUGCCAUUUGGGUAUUUGUGUGAUUUUUCUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUGGGGAAACUAAAAAACCGCACUUUG ((((..-.....(((((.....)))))..)))).....(((..((((............))))..)))......((((((((((((...(((....)))...)).))))))))))..... ( -26.20) >consensus GCAUUUUUCUCUAUGCCACU__GGUAUUUGGGUGAUUUUUCUUUUGAUUCUGCUGCAUUUCAAUUGAACUUUGUUGCGGUUUUUGGCUGCCCAUUUGGGAAACUAAAAAACCGCACUUUG ........(((.(((((.....)))))..)))......(((..((((............))))..)))......((((((((((((...(((....)))...)).))))))))))..... (-21.94 = -22.54 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:49 2006