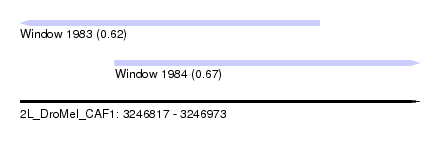
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,246,817 – 3,246,973 |
| Length | 156 |
| Max. P | 0.667291 |

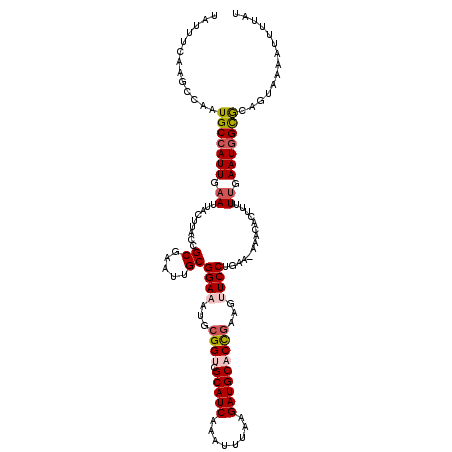
| Location | 3,246,817 – 3,246,934 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3246817 117 - 22407834 AACUUGGGUGCAUCUUAAAUUUGAUGCGACCGCAUUUCCGCAAUUCGCGGUAAGUAAUUCAAUGGCAUUGGCUUGAAAUAAAAUUUGAUGCCCCUCAUGUGGCAGGCAGUUGGUGGC--- .(((((((((((((........))))).)))......((((.....)))))))))........(.(((..(((..((......))..)((((((......))..))))))..))).)--- ( -32.40) >DroSec_CAF1 80890 113 - 1 AACUGCGGUGCAUCUUAAAUUUGAUGCGACCGCAUUUCCGCAAUUCGCGGUAAGUAAUUCAAUGGCAUUGGCUUGAAAUAAAAUUUGAUGCCCCUCAUGUGGC----AGUUGGUGGC--- ....((((((((((........))))).)))))((((((((.....)))).))))........(.(((..(((..((......))..)((((.(....).)))----)))..))).)--- ( -35.70) >DroSim_CAF1 84030 113 - 1 AACUGCGGUGCAUCUUAAAUUUGAUGCGACCGCAUUUCCGCAAUUCGCGGUAAGUAAUUCAAUGGCAUUGGCUUGAAAUAAAUUUUGAUGCCCCUCAUGUGGC----AGUUGGUGGC--- ....((((((((((........))))).)))))((((((((.....)))).))))........(.(((..(((..(((....)))..)((((.(....).)))----)))..))).)--- ( -36.40) >DroEre_CAF1 77677 111 - 1 AACUUCAGAGCAUCUUAAAUUUGAUGCGACCGCGCAUCCGCAAUUCGCAGUAUGUAAUUCAAUGGCAGUGGCUUCAAAUAAAAUUUGAUGCCC--CCACUGGC----AGUUGGUGGA--- .........(((((........)))))..(((((((((.((.....)).).))))....(((((.(((((((.((((((...)))))).))).--..)))).)----).))))))).--- ( -32.30) >DroYak_CAF1 76914 116 - 1 AACUUCGGAGCAUCUUAAAUUUGAUGCGACCGCAUAUCCGCAAUUCGCAGAAUGUAAUUCAAUUGCAGUGGCUUCAAAUAAAAUUUAAUGCCUCUCCACUGGC----AGUUGGCGGAUAU .....(((.(((((........)))))..))).((((((((.((((...))))......((((((((((((......((........))......))))).))----))))))))))))) ( -35.10) >consensus AACUUCGGUGCAUCUUAAAUUUGAUGCGACCGCAUUUCCGCAAUUCGCGGUAAGUAAUUCAAUGGCAUUGGCUUGAAAUAAAAUUUGAUGCCCCUCAUGUGGC____AGUUGGUGGC___ .........(((((........)))))..((((((((((((.....)))).))))....((((((((((((.((.......)).))))))))((......))......)))))))).... (-23.58 = -23.70 + 0.12)

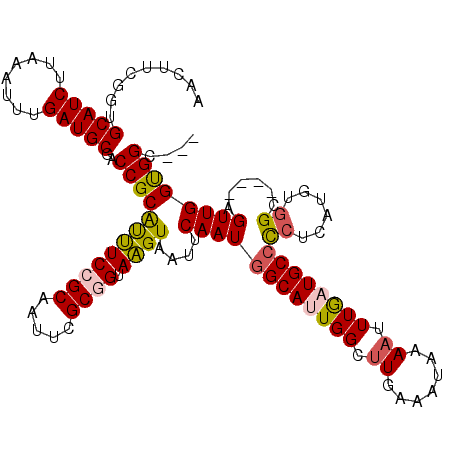
| Location | 3,246,854 – 3,246,973 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -21.56 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3246854 119 + 22407834 UAUUUCAAGCCAAUGCCAUUGAAUUACUUACCGCGAAUUGCGGAAAUGCGGUCGCAUCAAAUUUAAGAUGCACCCAAGUUCCUGAA-AAACACUUUUUGAAUGGCACAGUAAAAUUUUAU .............((((((((((((.....((((.....))))...((.(((.(((((........)))))))))))))))..(((-((....))))).))))))).............. ( -27.60) >DroSec_CAF1 80923 120 + 1 UAUUUCAAGCCAAUGCCAUUGAAUUACUUACCGCGAAUUGCGGAAAUGCGGUCGCAUCAAAUUUAAGAUGCACCGCAGUUCCUGAAAAAACACUUUUUGAAUGGUGCAGUAAAAUUUUAU ...(((((((....))..)))))(((((((((((.....))((((.((((((.(((((........))))))))))).))))....................)))).)))))........ ( -34.00) >DroSim_CAF1 84063 119 + 1 UAUUUCAAGCCAAUGCCAUUGAAUUACUUACCGCGAAUUGCGGAAAUGCGGUCGCAUCAAAUUUAAGAUGCACCGCAGUUCCUGAA-AAACACUUUUUGAAUGGCGCAGUAAAAUUUUAU ..............((((((.((.........((.....))((((.((((((.(((((........))))))))))).))))....-.........)).))))))............... ( -33.40) >DroEre_CAF1 77708 119 + 1 UAUUUGAAGCCACUGCCAUUGAAUUACAUACUGCGAAUUGCGGAUGCGCGGUCGCAUCAAAUUUAAGAUGCUCUGAAGUUCCUGGA-AAGCGCUUUUUGAAUGGCGCAAUAAAAUUUUAC ..............((((((.((.......((((.....))))..((((..(((((((........)))))...(.....)...))-..))))...)).))))))............... ( -27.30) >DroYak_CAF1 76950 119 + 1 UAUUUGAAGCCACUGCAAUUGAAUUACAUUCUGCGAAUUGCGGAUAUGCGGUCGCAUCAAAUUUAAGAUGCUCCGAAGUUCCUGAA-AAACACUUUUCGAAUGGCGCACUAAAAUUUUAU ........((((..((((((((((...))))....))))))(((....(((..(((((........))))).)))....)))((((-((....))))))..))))............... ( -26.90) >consensus UAUUUCAAGCCAAUGCCAUUGAAUUACUUACCGCGAAUUGCGGAAAUGCGGUCGCAUCAAAUUUAAGAUGCACCGAAGUUCCUGAA_AAACACUUUUUGAAUGGCGCAGUAAAAUUUUAU .............(((((((.((.........((.....))((((...((((.(((((........)))))))))...))))..............)).))))))).............. (-21.56 = -22.48 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:44 2006