
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,245,870 – 3,246,190 |
| Length | 320 |
| Max. P | 0.999807 |

| Location | 3,245,870 – 3,245,990 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -38.62 |
| Energy contribution | -40.00 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245870 120 + 22407834 AAAACAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUCUCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGA ....((((.....((((.(((((..(((((((.......)))))))..))....)))))))........((((((((((((....))))))))))))))))................... ( -42.20) >DroSec_CAF1 79938 120 + 1 AAAACAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGA ....((((.((((((((.(((((..(((((((.......)))))))..))....))))))).)))).))))(((.(((((((((....))))))))).)))................... ( -45.40) >DroSim_CAF1 83063 120 + 1 AAAACAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGA ....((((.((((((((.(((((..(((((((.......)))))))..))....))))))).)))).))))(((.(((((((((....))))))))).)))................... ( -45.40) >DroYak_CAF1 75938 120 + 1 AAAAGAGAACAGCGCAGUACAGAAAGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUUUCACUACAACUGCAGCUGCAACUGCAACUGUAGCUGCAUCUGAAUUCAAAUACGU ....(((..(((.(((((((((...(((((((.......))))))).........(.((((((((.......)))))))))(((....))).)))).)))))..)))..)))........ ( -35.80) >consensus AAAACAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGA ....((((.((((((((.(((((..(((((((.......)))))))..))....))))))).)))).))))(((.(((((((((....))))))))).)))................... (-38.62 = -40.00 + 1.38)

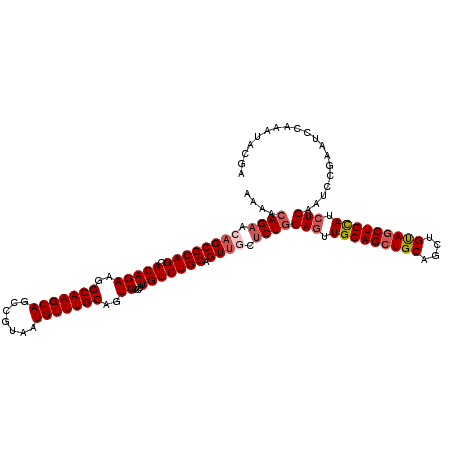

| Location | 3,245,870 – 3,245,990 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -37.61 |
| Energy contribution | -37.42 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245870 120 - 22407834 UCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGAGACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUGUUUU ..((((((((((....))))))))))(((.(((((.(((((.(((.((((((.....)))))).(((((.(.(((.........)))).))))).....)))))))))))))...))).. ( -41.90) >DroSec_CAF1 79938 120 - 1 UCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGCAACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUGUUUU ..((((((((((....))))))))))(((.(((((.(((((.(((..(((((..(((((.......))))).)))))........(((.....)))...)))))))))))))...))).. ( -43.70) >DroSim_CAF1 83063 120 - 1 UCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGCAACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUGUUUU ..((((((((((....))))))))))(((.(((((.(((((.(((..(((((..(((((.......))))).)))))........(((.....)))...)))))))))))))...))).. ( -43.70) >DroYak_CAF1 75938 120 - 1 ACGUAUUUGAAUUCAGAUGCAGCUACAGUUGCAGUUGCAGCUGCAGUUGUAGUGAAACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCUUUCUGUACUGCGCUGUUCUCUUUU ..(((((((....)))))))((..(((((.(((((.(((((.((((((((((((....(((((.(.....).)))))..))))))))))))..))....))))))))))))).))..... ( -44.20) >consensus UCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGAAACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUGUUUU ..((((((((((....))))))))))((..(((((.(((((.(((.((((((.....)))))).(((((.(.(((.........)))).))))).....))))))))))))).))..... (-37.61 = -37.42 + -0.19)



| Location | 3,245,910 – 3,246,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -24.69 |
| Energy contribution | -26.25 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245910 120 + 22407834 GUUUGCAGUCAACUUGUCUGCAGUCUCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGAGUACGAAUCCAAAUCUGCUGGAAUGCAAUUGUUGACAGUA ...(((.((((((.....(((((((((..((((((((((((....))))))))))))((.......))..........))).))...((((.......)))).))))...)))))).))) ( -38.00) >DroSec_CAF1 79978 120 + 1 GUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGAAUCCGAAUCCGAAUCUGCUGGAAUGCAAUUGUUGACAGUA ...........(((.(((.(((((((((((((((.(((((((((....)))))))))((.((.((.((.((.......)).)).)).)).))..)))).)))..)))))))).)))))). ( -40.70) >DroSim_CAF1 83103 120 + 1 GUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGAAUCCGAAUCCGAAUCUGCUGGAAUGCAAUUGUUGACAGUA ...........(((.(((.(((((((((((((((.(((((((((....)))))))))((.((.((.((.((.......)).)).)).)).))..)))).)))..)))))))).)))))). ( -40.70) >DroEre_CAF1 76804 108 + 1 GUUUGCAGUCAACUUGUCUGCAGUCCCUCU------------GCAGCUGCAUCUGAAUCCGAGUCUGAAUCCGAGUCCGAAUGCGAAUCUGAAUCUGCUGGAAUGCAAUUGUUGACAGUA ...(((.((((((..(.((((((.....))------------)))))(((((.....(((.(((..((.((.((...((....))..)).)).)).)))))))))))...)))))).))) ( -28.90) >DroYak_CAF1 75978 120 + 1 GUUUGCAGUCAACUUGUCUGCAGUUUCACUACAACUGCAGCUGCAACUGCAACUGUAGCUGCAUCUGAAUUCAAAUACGUAUCCGAAUCAGAAUCUGCUGAAAUGCAAUUGUUGACAGUA ...(((.((((((.....((((.(((((.......((((((((((........))))))))))(((((.(((............))))))))......)))))))))...)))))).))) ( -36.20) >consensus GUUUGCAGUCAACUUGUCUGCAGUUGCUCUGCAGUUGCAGCUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGAAUCCGAAUCCGAAUCUGCUGGAAUGCAAUUGUUGACAGUA ...(((.((((((.....((((.........(((.(((((((((....))))))))).)))........((((.........................)))).))))...)))))).))) (-24.69 = -26.25 + 1.56)


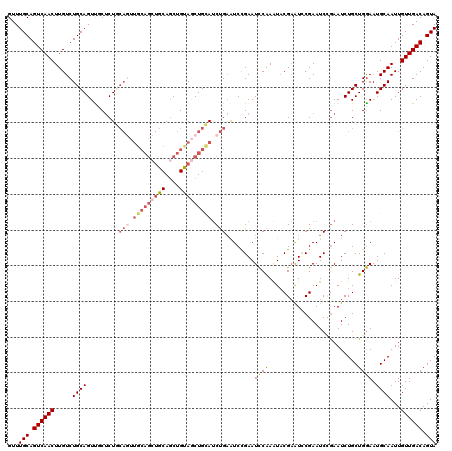
| Location | 3,245,910 – 3,246,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -28.58 |
| Energy contribution | -31.66 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245910 120 - 22407834 UACUGUCAACAAUUGCAUUCCAGCAGAUUUGGAUUCGUACUCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGAGACUGCAGACAAGUUGACUGCAAAC ....((((((............(((((((((((((((.(..((....))..).)))))))))))(((((....)))))..))))..((((((.....))))))....))))))....... ( -43.70) >DroSec_CAF1 79978 120 - 1 UACUGUCAACAAUUGCAUUCCAGCAGAUUCGGAUUCGGAUUCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGCAACUGCAGACAAGUUGACUGCAAAC ....((((((............((((....(((((((((((((....)))))))))))))...((((((....)))))).))))..((((((.....))))))....))))))....... ( -47.90) >DroSim_CAF1 83103 120 - 1 UACUGUCAACAAUUGCAUUCCAGCAGAUUCGGAUUCGGAUUCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGCAACUGCAGACAAGUUGACUGCAAAC ....((((((............((((....(((((((((((((....)))))))))))))...((((((....)))))).))))..((((((.....))))))....))))))....... ( -47.90) >DroEre_CAF1 76804 108 - 1 UACUGUCAACAAUUGCAUUCCAGCAGAUUCAGAUUCGCAUUCGGACUCGGAUUCAGACUCGGAUUCAGAUGCAGCUGC------------AGAGGGACUGCAGACAAGUUGACUGCAAAC ....((((((..((((......))))..........((((..(((..(((........)))..)))..)))).(((((------------((.....)))))).)..))))))....... ( -31.00) >DroYak_CAF1 75978 120 - 1 UACUGUCAACAAUUGCAUUUCAGCAGAUUCUGAUUCGGAUACGUAUUUGAAUUCAGAUGCAGCUACAGUUGCAGUUGCAGCUGCAGUUGUAGUGAAACUGCAGACAAGUUGACUGCAAAC ....((((((..(((((((((.(((...((((((((((((....))))))).)))))))).((((((..((((((....))))))..)))))))))).)))))....))))))....... ( -44.10) >consensus UACUGUCAACAAUUGCAUUCCAGCAGAUUCGGAUUCGGAUUCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAGCUGCAACUGCAGAGAAACUGCAGACAAGUUGACUGCAAAC ....((((((..((((......))))((((((((((((((....)))))))))))))).(((.((((((....)))))).)))...((((((.....))))))....))))))....... (-28.58 = -31.66 + 3.08)

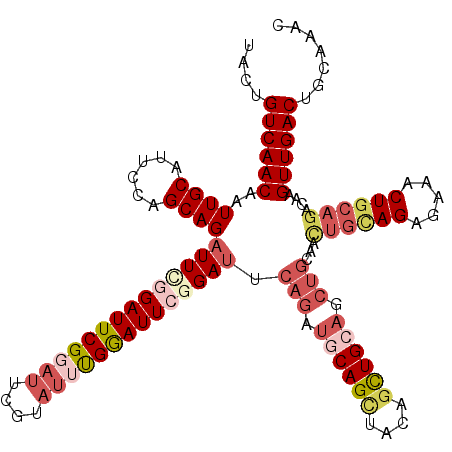
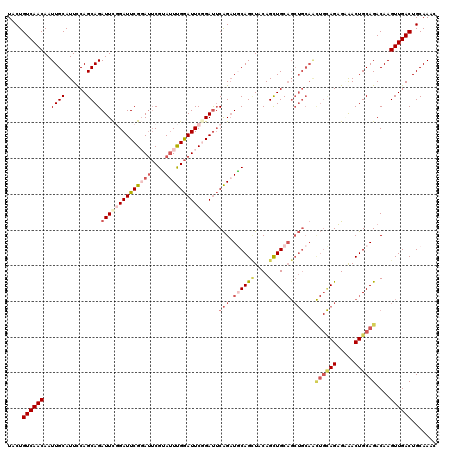
| Location | 3,246,030 – 3,246,150 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -37.10 |
| Energy contribution | -37.74 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3246030 120 + 22407834 CAAAAGAAGUGACAAAAGGUUCCGAGAGCGUUGCCUGUUCUAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCG ..........(((.....((((...))))(((((.((((((((((((((..(((...(((......)))....)))..)))))))))))))).))))).......(((....))).))). ( -40.90) >DroSec_CAF1 80098 120 + 1 CAAAAGAAGUGACAAAAGGAUCCGAGAGCGUUGCCAGUUCUAGUUGGCCACCUCCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCGAUUAUUUAGGCCACAUGGCGGUUG .....................((.(((..(((((..(((((((((((((..(((...(((......)))....)))..)))))))))))))..)))))..)))..(((....)))))... ( -37.80) >DroSim_CAF1 83223 120 + 1 CAAAAGAAGUGACAAAAGGUUCCGAGAGCGUUGCCAGUUCUAGUUGGCCACCUCCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCGAUUAUUUAGGCCACAUGGCGGUCG ..........(((....(((..((....))..))).(((((((((((((..(((...(((......)))....)))..)))))))))))))..............(((....))).))). ( -40.90) >DroEre_CAF1 76912 120 + 1 CAAAAGAAGUGACAAAAGGUUCCGAGUGCGUUGCCUGUUCUAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCG ..........(((........(..((((.(((((.((((((((((((((..(((...(((......)))....)))..)))))))))))))).)))))))))..)(((....))).))). ( -41.90) >DroYak_CAF1 76098 109 + 1 CAAAAGAAGUGACAAAAGGU-----------CGCCUGUGCUAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCG .....(((((((...(((((-----------.(((..........))).))))).)))))))....((((.(((.((.(((((((((.......))))).....)))))).))).)))). ( -36.10) >consensus CAAAAGAAGUGACAAAAGGUUCCGAGAGCGUUGCCUGUUCUAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCG ..........(((................(((((.((((((((((((((..(((...(((......)))....)))..)))))))))))))).))))).......(((....))).))). (-37.10 = -37.74 + 0.64)



| Location | 3,246,070 – 3,246,190 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -38.36 |
| Energy contribution | -37.68 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3246070 120 + 22407834 UAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCGCCAGCAGGGCAAGGUUAACUGCCAACCGCCAAUGGGUUAG (((((((((..(((...(((......)))....)))..))))))))).................((((.(((((((((.(((.....)))..(((.....))).)))))).))))))).. ( -41.60) >DroSec_CAF1 80138 120 + 1 UAGUUGGCCACCUCCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCGAUUAUUUAGGCCACAUGGCGGUUGCCAGCACGGCAAGGUUAACUGCCAACCGCCAAUGGAUUAG (((((((((......(((((((((.(((((........))))).)))))....)))).......)))).(((((((((((((.....)))..(((.....)))))))))).)))))))). ( -44.62) >DroSim_CAF1 83263 120 + 1 UAGUUGGCCACCUCCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCGAUUAUUUAGGCCACAUGGCGGUCGCCAGCACGGCAAUGUUAACUGCCAACCGCCAAUGGAUUAG ..((((((.(((...(((((((((.(((((........))))).)))))....))))........(((....)))))).))))))..((((........))))..(((....)))..... ( -40.50) >DroEre_CAF1 76952 120 + 1 UAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCGCUAGCCCGGCAAGGUUAACUGCCAACCGCCAAUGCGUUGG (((((((((.................((((.(((.((.(((((((((.......))))).....)))))).))).))))(((.....)))..)))))))))(((((.((....))))))) ( -38.40) >DroYak_CAF1 76127 120 + 1 UAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCGCUGGCCCAGCAAGGUUAACUGCCAACCGCCAAUGGGUUAG (((((((((..(((...(((......)))....)))..))))))))).................((((.(((((((((.((((...))))..(((.....))).)))))).))))))).. ( -41.00) >consensus UAGUUGGCCACCUUCUCGCUUUAACUGGCCACCGAGUAGGCCAAUUGGAACGUGCAAUUAUUUAGGCCACAUGGCGGUCGCCAGCACGGCAAGGUUAACUGCCAACCGCCAAUGGGUUAG ....(((((...(((.((.(((((.(((((........))))).))))).)).).)).......)))))(((((((((.(((.....)))..(((.....))).)))))).)))...... (-38.36 = -37.68 + -0.68)



| Location | 3,246,070 – 3,246,190 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -45.86 |
| Consensus MFE | -43.94 |
| Energy contribution | -43.90 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3246070 120 - 22407834 CUAACCCAUUGGCGGUUGGCAGUUAACCUUGCCCUGCUGGCGACCGCCAUGUGGCCUAAAUAAUUGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGAAGGUGGCCAACUA .........(((((((((.((((............)))).)))))))))..(((((..........(.((((..((((((((........))))))))..)))).).....))))).... ( -45.76) >DroSec_CAF1 80138 120 - 1 CUAAUCCAUUGGCGGUUGGCAGUUAACCUUGCCGUGCUGGCAACCGCCAUGUGGCCUAAAUAAUCGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGGAGGUGGCCAACUA .........(((((((((.((((............)))).)))))))))..(((((.......((.(.((((..((((((((........))))))))..)))).)))...))))).... ( -48.20) >DroSim_CAF1 83263 120 - 1 CUAAUCCAUUGGCGGUUGGCAGUUAACAUUGCCGUGCUGGCGACCGCCAUGUGGCCUAAAUAAUCGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGGAGGUGGCCAACUA .........(((((((((.((((..((......)))))).)))))))))..(((((.......((.(.((((..((((((((........))))))))..)))).)))...))))).... ( -48.30) >DroEre_CAF1 76952 120 - 1 CCAACGCAUUGGCGGUUGGCAGUUAACCUUGCCGGGCUAGCGACCGCCAUGUGGCCUAAAUAAUUGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGAAGGUGGCCAACUA .....((((.((((((((((((......)))))..(....))))))))))))((((..........(.((((..((((((((........))))))))..)))).).....))))..... ( -43.06) >DroYak_CAF1 76127 120 - 1 CUAACCCAUUGGCGGUUGGCAGUUAACCUUGCUGGGCCAGCGACCGCCAUGUGGCCUAAAUAAUUGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGAAGGUGGCCAACUA .............(((((((.....(((((..(((((((((....))....)))))))..........((((..((((((((........))))))))..))))..))))).))))))). ( -44.00) >consensus CUAACCCAUUGGCGGUUGGCAGUUAACCUUGCCGUGCUGGCGACCGCCAUGUGGCCUAAAUAAUUGCACGUUCCAAUUGGCCUACUCGGUGGCCAGUUAAAGCGAGAAGGUGGCCAACUA .........(((((((((.((((............)))).)))))))))..(((((.......((.(.((((..((((((((........))))))))..)))).)))...))))).... (-43.94 = -43.90 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:41 2006