
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,244,940 – 3,245,170 |
| Length | 230 |
| Max. P | 0.974521 |

| Location | 3,244,940 – 3,245,054 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -20.11 |
| Energy contribution | -21.23 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3244940 114 + 22407834 UAUAUCUAUAUCUACAUGAGCUCUCUCUCUCUACAUAUAUAU------AUGUUAAGCCUGCGGACAGACGACCUGAAGUAGAUGCAAUGAACAGGGGAGCUUUAUGGGUAUUGUAGGUAA ..((((((((....((((((((((((...((.(((((....)------))))...((((((...(((.....)))..))))..))...))...))))))).))))).....)))))))). ( -30.00) >DroSec_CAF1 78999 120 + 1 UAUAUCUAUAUCUACAUGAGCUCUCACUACAUAUAUAUGUAUAUGUAUGUGUAAAGCCUGCGGACAGACGACCUGACGUAGAUGCAAUGAACAGGGGAGCUUUAUAGGUAUUGUAGGUAA ..((((((((((((...((((((((..(((((((((((....)))))))))))..(((((((..(((.....))).)))))..)).........))))))))..)))))...))))))). ( -39.20) >DroSim_CAF1 82129 116 + 1 UAUAUCUAUAUCUACAUGAGCUCUCACUACAU----AUGUAUAUCUAUGUGUAAAGCCUGCGGACAGACGACCUGGAGUAGAUGCAAUGAACAGGGGAGCUUUAUGGGUAUUGUAGGUAA ..((((((((....(((((((((((..(((((----(((......))))))))..((((((...(((.....)))..))))..)).........)))))).))))).....)))))))). ( -34.70) >DroEre_CAF1 75875 106 + 1 UAUAUCUAUAUCUACAUGAGCUCUCACAAUAU--------------AUGUGUAAAGCCUGCGGACAGACGAGCUGAAGUGGAUGCAAUGAACAGGGGAGCUUUAUGGGUAUUGUAGGUAA ..(((((((((((((.(.(((((.((((....--------------.))))....(.(((....))).)))))).).)))))))(((((..((((....))...))..))))))))))). ( -27.50) >DroYak_CAF1 75045 102 + 1 UAUAUCUAUAUCUACACGAGCUCUCACAAUAUACAU----ACUCGUAUGUGUAAAGCCGGCGGACAG--------------AUGCAAUGAACAGGGGAGCUUUAUGGGUAUUGUAGGUAA ..((((((((.((....((((((((....(((((((----(....))))))))......(((.....--------------.))).........))))))))....))...)))))))). ( -29.00) >consensus UAUAUCUAUAUCUACAUGAGCUCUCACUAUAUA_AUAU_UAU___UAUGUGUAAAGCCUGCGGACAGACGACCUGAAGUAGAUGCAAUGAACAGGGGAGCUUUAUGGGUAUUGUAGGUAA ..((((((((((((...((((((((...((((................))))...((((((...(((.....)))..))))..)).........))))))))..)))))...))))))). (-20.11 = -21.23 + 1.12)


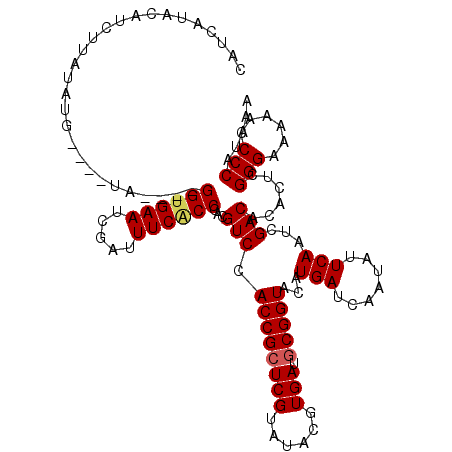
| Location | 3,245,054 – 3,245,170 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -28.78 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245054 116 + 22407834 UUUAUGGGGUUUUUCCCGAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGAGCGGUGGACGUGGGUGAAAUCGAUUCACCUACCUA----CAUAUAAGAUGUAUGAUG ....(((((....))))).....((((((..(...)..)))...((((((.((........)))))))))))(((((((((.....)))))))))...----(((((.....)))))... ( -35.00) >DroSec_CAF1 79119 112 + 1 UUUAUGGGGUUUUUCCCGAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGAGCGGUGGACGUGGGUGAAAUCGAUUCACC----UA----CAUAUAAGAUGUAUGAUG ....(((((....))))).(((..(((((..(...)..))).(.((((((.((........)))))))).).(((((((((.....))))))----))----)......))..))).... ( -34.60) >DroSim_CAF1 82245 112 + 1 UUUAUGGGGUUUUUCCCGAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGAGCGGUGGACGUGGGUGAAAUCGAUUCGCC----UA----CAUAUAAGAUGUAUGAUG ....(((((....))))).(((..(((((..(...)..))).(.((((((.((........)))))))).).(((((((((.....))))))----))----)......))..))).... ( -34.20) >DroEre_CAF1 75981 115 + 1 UUUAUGGGGUUU-UCCCCAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGAGCGGUGGACGUGGGCGAAAUCGAUUCACC----UAGGAGUAUAUAAGAUGUAUGAUG ....(((((...-.)))))(((.(((((((.....(((..((((((((((.((........)))))))..)))))..)))))))))).))).----.....((((((...)))))).... ( -33.60) >DroYak_CAF1 75147 112 + 1 UUUAUGGGGUUUUUCCCGAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGACCGGUGGACGUGGGUGAAAUCGAUUCACC----UA----CAUAUAAGAUGUAUGCUG ....(((((....))))).(((..(((((..(...)..)))..((.((((....((....))....))))))(((((((((.....))))))----))----)......))..))).... ( -32.20) >consensus UUUAUGGGGUUUUUCCCGAGUGUGUCGAUUGAAUAUUGAUCAUGUACCGCAUCACGUAUACGAGCGGUGGACGUGGGUGAAAUCGAUUCACC____UA____CAUAUAAGAUGUAUGAUG .....((((....))))..(((.(((((((.........((((((.((((....((....))....))))))))))....))))))).)))...........(((((.....)))))... (-28.78 = -28.46 + -0.32)

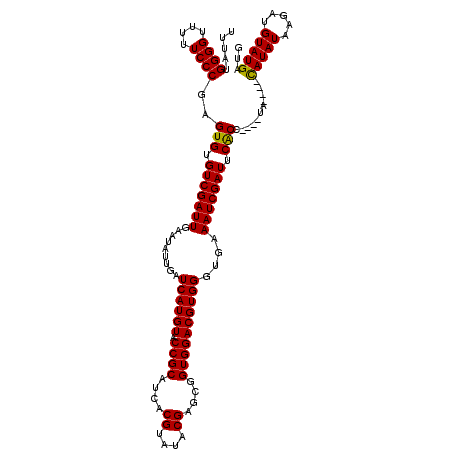

| Location | 3,245,054 – 3,245,170 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3245054 116 - 22407834 CAUCAUACAUCUUAUAUG----UAGGUAGGUGAAUCGAUUUCACCCACGUCCACCGCUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUCGGGAAAAACCCCAUAAA .....(((((.....)))----)).((.((((((.....)))))).))(((.((((((((......))).)))))...(((.......)))...)))......(((......)))..... ( -27.90) >DroSec_CAF1 79119 112 - 1 CAUCAUACAUCUUAUAUG----UA----GGUGAAUCGAUUUCACCCACGUCCACCGCUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUCGGGAAAAACCCCAUAAA .(((((.........(((----(.----((((((.....)))))).))))..((((((((......))).)))))..))))).....................(((......)))..... ( -26.20) >DroSim_CAF1 82245 112 - 1 CAUCAUACAUCUUAUAUG----UA----GGCGAAUCGAUUUCACCCACGUCCACCGCUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUCGGGAAAAACCCCAUAAA .(((((.........(((----(.----((.(((.....))).)).))))..((((((((......))).)))))..))))).....................(((......)))..... ( -22.30) >DroEre_CAF1 75981 115 - 1 CAUCAUACAUCUUAUAUACUCCUA----GGUGAAUCGAUUUCGCCCACGUCCACCGCUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUGGGGA-AAACCCCAUAAA ........................----((((((.....))))))...(((.((((((((......))).)))))...(((.......)))...)))....(((((.-...))))).... ( -27.00) >DroYak_CAF1 75147 112 - 1 CAGCAUACAUCUUAUAUG----UA----GGUGAAUCGAUUUCACCCACGUCCACCGGUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUCGGGAAAAACCCCAUAAA ..(.(((((((....(((----(.----((((((.....)))))).)))).....))).)))).)(((.((((((...(((.......))))))).)))))..(((......)))..... ( -24.70) >consensus CAUCAUACAUCUUAUAUG____UA____GGUGAAUCGAUUUCACCCACGUCCACCGCUCGUAUACGUGAUGCGGUACAUGAUCAAUAUUCAAUCGACACACUCGGGAAAAACCCCAUAAA ............................((((((.....))))))...(((.((((((((......))).)))))...(((.......)))...)))......(((......)))..... (-21.86 = -22.14 + 0.28)

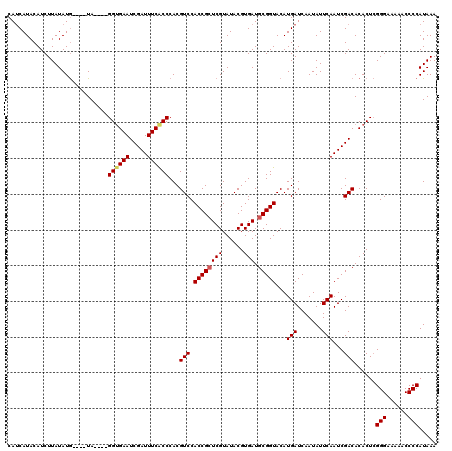
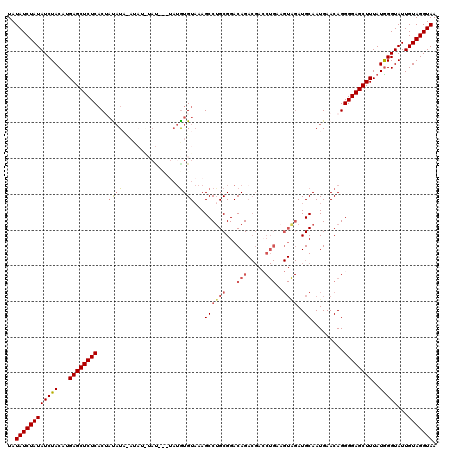
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:35 2006