
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,244,140 – 3,244,336 |
| Length | 196 |
| Max. P | 0.999514 |

| Location | 3,244,140 – 3,244,256 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.26 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -41.84 |
| Energy contribution | -42.44 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
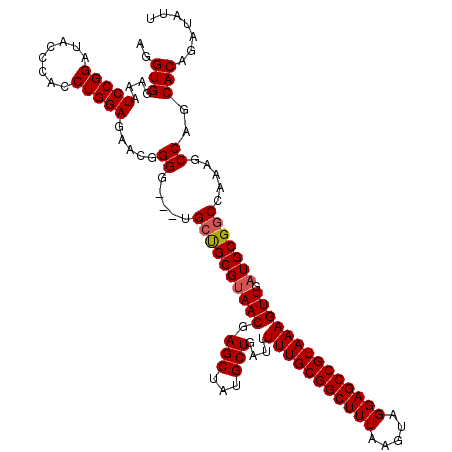
>2L_DroMel_CAF1 3244140 116 + 22407834 CUUAAACCGUUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGCCGGCAUACGCUGCGGCU-CAGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UG ......((((((.(....).(((((((..(((((....)))))((((((((.....)))).))))......)))))))-(((((((..((....))..)))))))..))))))..---.. ( -46.10) >DroSec_CAF1 78198 116 + 1 CUUAAACCGUUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGGCGGCAUACGCUGCGGCC-CAGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UG ......((((((.(....)..(((((...(((((....)))))(((((((.....)))))))(((....))))))))(-(((((((..((....))..)))))))).))))))..---.. ( -49.10) >DroSim_CAF1 81340 116 + 1 CUUAAACCGUUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGGCGGCAUACGCUGCGGCC-CAGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UG ......((((((.(....)..(((((...(((((....)))))(((((((.....)))))))(((....))))))))(-(((((((..((....))..)))))))).))))))..---.. ( -49.10) >DroEre_CAF1 75131 116 + 1 CUUAAACCGCUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGGCGGCAUACGCUGCGGCC-CAGGUGGAAAUCCGGAUUACCACCUGGAGAACGGGG---UG ......(((((..(....)..(((((.....)).))).)))))(((.(((.......((.(((((....))))).))(-(((((((.(((....))).))))))))...))).))---). ( -47.20) >DroYak_CAF1 74249 120 + 1 CUUAAACCGUUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGGCGGCAUACGCUGCGGCCCCACGUGGAAAUCCGGAUACCCACCUGGAUAACGGGGGACUG ......((((...(....).(((.(((..(((((....)))))(((((((.....)))))))....))))))))))((((.(((....((((((........)))))).))))))).... ( -43.10) >consensus CUUAAACCGUUCAGGAAACAAGCUGUAAAUCGCUGGCAAGCGGGCCGUGUAAUAAAUGCGGCGGCAUACGCUGCGGCC_CAGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG___UG ......((((((.(....)..((((((..(((((....)))))((((((((.....)))).))))......))))))..(((((((..((....))..)))))))..))))))....... (-41.84 = -42.44 + 0.60)



| Location | 3,244,219 – 3,244,336 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3244219 117 + 22407834 AGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UGCUGCGUAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUAGGAGCCGUAAAGUUGAUGCGGCCAAAGCCAGCACAGAUAUU ((((((..((....))..))))))..........(---(((((((((((.(((...)))....(((((((((((.....)))))))))))))).))))(((....))))))))....... ( -39.50) >DroSec_CAF1 78277 117 + 1 AGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UGCUGCGUAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUGGGAGCCGUAAAGUUGAUGCGGCCAAAGCCGGCACAGAUAUU ((((((..((....))..))))))..........(---(((((((((((.(((...)))....(((((((((((.....)))))))))))))).))))(((....))))))))....... ( -39.60) >DroSim_CAF1 81419 117 + 1 AGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG---UGCUGCGUAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUAGGAGCCGUAAAGUUGAUGCGACCAAAGCCGGCACAGAUAUU ..(((.....((((.....(((((.(....).)))---)).((((((((.(((...)))....(((((((((((.....)))))))))))))).))))).......)))))))....... ( -34.60) >DroEre_CAF1 75210 117 + 1 AGGUGGAAAUCCGGAUUACCACCUGGAGAACGGGG---UGCCGCGCAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUAGGAGCCGUAAAGUUGAUGCGGCCAAAGCCCGCACAGAUAUU ((((((.(((....))).))))))......((((.---(((((((((((.(((...)))....(((((((((((.....))))))))))))))).))))))...).)))).......... ( -46.10) >DroYak_CAF1 74329 120 + 1 ACGUGGAAAUCCGGAUACCCACCUGGAUAACGGGGGACUGCCGCGUAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUAGGAGCCGUAAAGUUGAUGCGGCCAAAGCCAGCACAGAUAUU ((((((...(((((....)).((((.....)))))))...)))))).....(((..((....((((((((((((.....)))))))))))).....))(((....))))))......... ( -43.90) >consensus AGGUGGAAAUCCGGAUACCCACCUGGAGAACGGGG___UGCUGCGUAACGAGCUAUGCUGAUUUUUGCGGCUUUAAGUAGGAGCCGUAAAGUUGAUGCGGCCAAAGCCAGCACAGAUAUU ..(((....(((((........))))).....((.....((((((((((.(((...)))....(((((((((((.....)))))))))))))).))))))).....))..)))....... (-35.14 = -35.30 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:31 2006