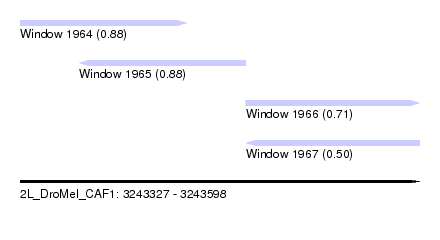
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,243,327 – 3,243,598 |
| Length | 271 |
| Max. P | 0.878020 |

| Location | 3,243,327 – 3,243,440 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.55 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3243327 113 + 22407834 AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCGCAAUCGGCGACUAAGCGCCGGAUUCCGAUUAGUAUUC------GGAUUCGGAUA-CGGAUUCCUCUUCAGAA ..((((...............((((((((..((((........))))....((((((......)))))).....))))))))...------(((((((....-))))))).))))..... ( -28.50) >DroSec_CAF1 77419 102 + 1 AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCGCAAUCGGCGACUAAACGCCGGAAUCCGAUU------------------CGAAUACCGGAUUCCUCUUCAGAA ..................(((((.......((((........(((((.......)))))....))))((((((((...------------------.......))))))))....))))) ( -26.50) >DroSim_CAF1 80549 102 + 1 AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCACAAUCGGCGACUAAACGCCGGAAUCCGAUU------------------CGAAUACCGGAUUCCUCUUCAGAA ..................(((((.......((((........(((((.......)))))....))))((((((((...------------------.......))))))))....))))) ( -26.30) >DroEre_CAF1 74363 107 + 1 AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCGCAAUCUGCGACUAAGCGCCGGAAUCCGAUUCCUAUU------------CGGAUU-CGUAUUCCUCUUGAAAA ..((((.((.....................((((........((((((.....))))))....)))).((((((((.......)------------))))))-).)).))))........ ( -27.90) >DroYak_CAF1 73454 119 + 1 AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCGCAAUCUGCGACUAAGCGCCGGAAUCCGAUUCCUAUUCGAAUACGGAUUCGGAUU-CGGAUUCCUCUUGAGUA ................(((((.........((((........((((((.....))))))....))))(((((((((.(((.(((((....)))))..))).)-))))))))....))))) ( -38.00) >consensus AAAGGAUUAAAUUAAAUAUUCUGCUAAUCAGGCGAAAGAAUUGUUGCGCAAUCGGCGACUAAGCGCCGGAAUCCGAUU__UAUU____________CGGAUA_CGGAUUCCUCUUCAGAA ....(((((...............))))).((((........(((((.......)))))....))))((((((((............................))))))))......... (-21.15 = -21.55 + 0.40)

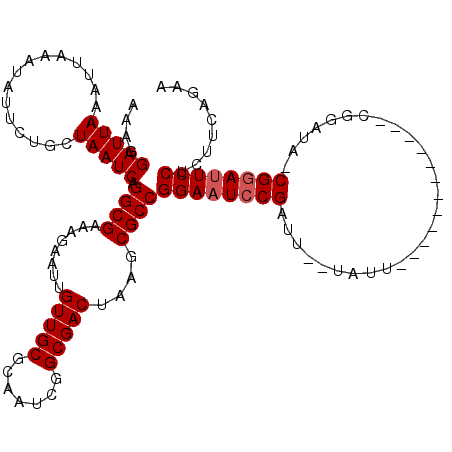
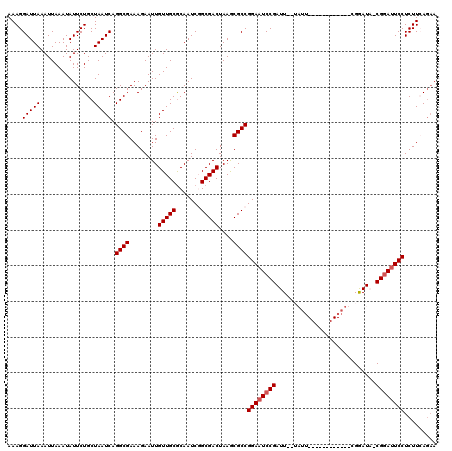
| Location | 3,243,367 – 3,243,480 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3243367 113 - 22407834 CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGCAUUCUGAAGAGGAAUCCG-UAUCCGAAUCC------GAAUACUAAUCGGAAUCCGGCGCUUAGUCGCCGAUUGCGCAACAA .....(((((((((...((.((.((((....)))).)))).........(((.((((-(((.((....)------).)))).....))).)))((((......)))))))))).)))... ( -29.60) >DroSec_CAF1 77459 102 - 1 CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGCAUUCUGAAGAGGAAUCCGGUAUUCG------------------AAUCGGAUUCCGGCGUUUAGUCGCCGAUUGCGCAACAA .....(((((((((...((.((.((((....)))).)))).........((((((((((.....------------------.))))))))))((((......)))))))))).)))... ( -36.90) >DroSim_CAF1 80589 102 - 1 CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGCAUUCUGAAGAGGAAUCCGGUAUUCG------------------AAUCGGAUUCCGGCGUUUAGUCGCCGAUUGUGCAACAA ..((((((((..(((..((.((.((((....)))).))))...)))...((((((((((.....------------------.))))))))))(((.....)))))))))))........ ( -33.70) >DroEre_CAF1 74403 107 - 1 CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGCAUUUUCAAGAGGAAUACG-AAUCCG------------AAUAGGAAUCGGAUUCCGGCGCUUAGUCGCAGAUUGCGCAACAA .....(((((((((.(((((((.((((....)))).)).....))))).(((((.((-(.(((.------------....))).))).)))))(((.....)))...)))))).)))... ( -30.50) >DroYak_CAF1 73494 119 - 1 CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGUAUACUCAAGAGGAAUCCG-AAUCCGAAUCCGUAUUCGAAUAGGAAUCGGAUUCCGGCGCUUAGUCGCAGAUUGCGCAACAA .....(((((((((...((((((.((.(.(((.(((......))).)))((((((((-(.(((..((.((....)).)).))).)))))))))).))..))))))..)))))).)))... ( -40.20) >consensus CUUAAUUGGUAAUCAUUUGACUGUCGACAUCUCGAGGGCAUUCUGAAGAGGAAUCCG_UAUCCG____________AAUA__AAUCGGAUUCCGGCGCUUAGUCGCCGAUUGCGCAACAA .....(((((((((...((.((.((((....)))).)))).........((((((((............................))))))))((((......)))))))))).)))... (-25.27 = -25.75 + 0.48)

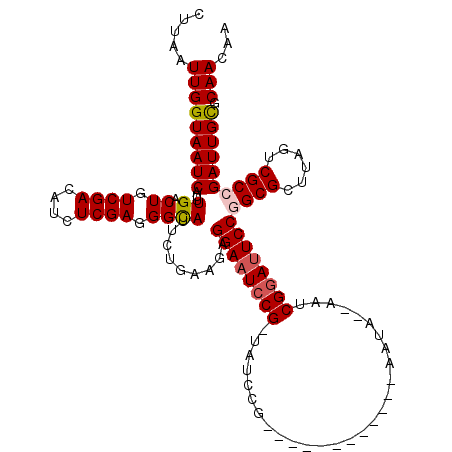
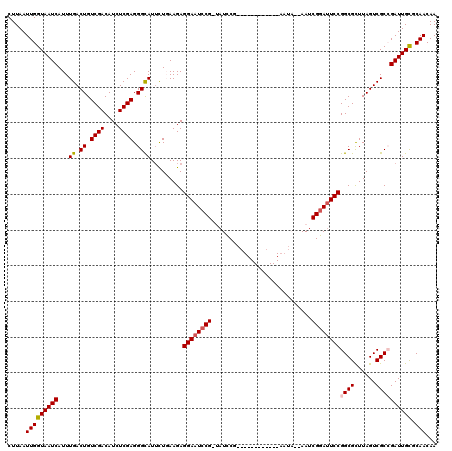
| Location | 3,243,480 – 3,243,598 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -20.24 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3243480 118 + 22407834 UAUGCAAAUUGUCUCUGGGCCACACAGACUCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGGGCUUUAUGGCGCAUUUUU--CUCCUCCUUAUCUAAUUCUAAUUGAGGACAGCACUUUU ..(((.....(((...(((((.((..((...((((.(((.....)))))))....))..)))))))...)))........--....((((((..((....))..))))))..)))..... ( -27.10) >DroSec_CAF1 77561 105 + 1 UAUGCAAAUUGUCUCUGGGCCACACAGACCCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGGGCUU-------------U--CUUCUCCUUAUCUAAAUCUAAUUGAGGACAGCACUUUU ..(((...........(((((.((..((...((((.(((.....)))))))....))..)))))))-------------.--....((((((..((....))..))))))..)))..... ( -22.50) >DroSim_CAF1 80691 118 + 1 UAUGCAAAUUGUCUCUGGGCCACACAGACCCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGCGCUUUAUGGCGCAUUUUU--UUUCUCCUUAUCUAAUUCUAAUUGAGGACAGCACUUUU ..(((.....((((.((.....)).))))..((((.(((.....)))))))........((((((....)))))).....--....((((((..((....))..))))))..)))..... ( -28.70) >DroEre_CAF1 74510 118 + 1 UAUGCAAAUUGUCUCUGUGCCACACGGACUCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGUGCUUAAUGGCGCAUUUUU--CUUCUCUUUAUCUAAUUCUAAUUGAGGACAGCACUUUU ..(((....((((((((((...))))))(((((((.(((.....)))))))........((((((....)))))).....--.......................))))))))))..... ( -27.90) >DroYak_CAF1 73613 120 + 1 UAUGCAAAUUGUCUCUGUGCCACACAGACUCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGGGCUUUGUGGCGCAUUUUUUGCUUCUCCUUAUCUAAUUCUAAUUGAGGACAGCACUUUU ...............(((((((((.((.(((((((.(((.....))))))).........))))).))))))))).....((((..((((((..((....))..)))))).))))..... ( -33.70) >consensus UAUGCAAAUUGUCUCUGGGCCACACAGACUCGCCAAGAUGUUCAAUCUGGCUUAAUCCUUGGGCUUUAUGGCGCAUUUUU__CUUCUCCUUAUCUAAUUCUAAUUGAGGACAGCACUUUU ..(((.....((((.((.....)).)))).(((((((((.....))))((((((.....))))))...))))).............((((((..((....))..))))))..)))..... (-20.24 = -21.32 + 1.08)

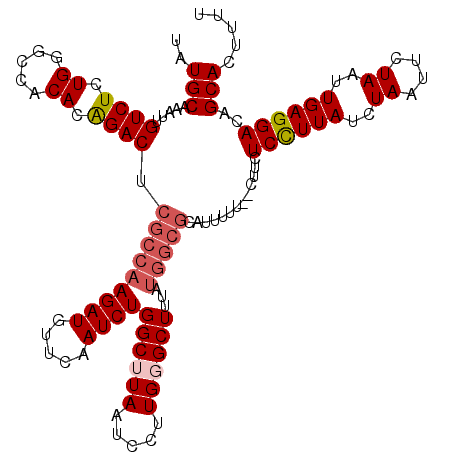
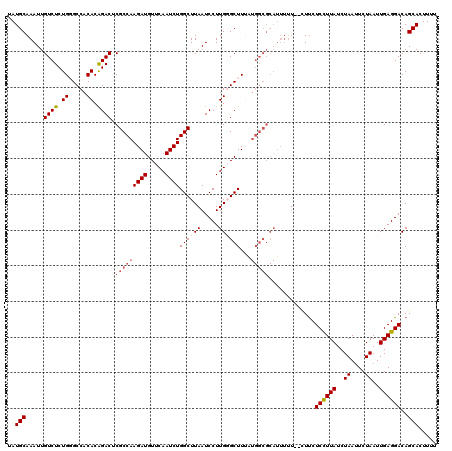
| Location | 3,243,480 – 3,243,598 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3243480 118 - 22407834 AAAAGUGCUGUCCUCAAUUAGAAUUAGAUAAGGAGGAG--AAAAAUGCGCCAUAAAGCCCAAGGAUUAAGCCAGAUUGAACAUCUUGGCGAGUCUGUGUGGCCCAGAGACAAUUUGCAUA ....(((((.(((((..(((........))).))))).--)....((.((((((........(((((..(((((((.....))).)))).))))).)))))).))..........)))). ( -28.10) >DroSec_CAF1 77561 105 - 1 AAAAGUGCUGUCCUCAAUUAGAUUUAGAUAAGGAGAAG--A-------------AAGCCCAAGGAUUAAGCCAGAUUGAACAUCUUGGCGGGUCUGUGUGGCCCAGAGACAAUUUGCAUA ....(((((((((((..(((........))).)))...--.-------------..(((((.((((...(((((((.....))).))))..)))).)).))).....))))....)))). ( -24.20) >DroSim_CAF1 80691 118 - 1 AAAAGUGCUGUCCUCAAUUAGAAUUAGAUAAGGAGAAA--AAAAAUGCGCCAUAAAGCGCAAGGAUUAAGCCAGAUUGAACAUCUUGGCGGGUCUGUGUGGCCCAGAGACAAUUUGCAUA ....((((((((.((.......................--.....(((((......)))))........(((((((.....))).))))(((((.....))))).))))))....)))). ( -31.10) >DroEre_CAF1 74510 118 - 1 AAAAGUGCUGUCCUCAAUUAGAAUUAGAUAAAGAGAAG--AAAAAUGCGCCAUUAAGCACAAGGAUUAAGCCAGAUUGAACAUCUUGGCGAGUCCGUGUGGCACAGAGACAAUUUGCAUA ....(((((((((((..(((........))).)))...--.....((.(((.....((((..(((((..(((((((.....))).)))).)))))))))))).))..))))....)))). ( -26.40) >DroYak_CAF1 73613 120 - 1 AAAAGUGCUGUCCUCAAUUAGAAUUAGAUAAGGAGAAGCAAAAAAUGCGCCACAAAGCCCAAGGAUUAAGCCAGAUUGAACAUCUUGGCGAGUCUGUGUGGCACAGAGACAAUUUGCAUA ....((((((((.((................((....(((.....))).)).....(((((.(((((..(((((((.....))).)))).))))).)).)))...))))))....)))). ( -29.90) >consensus AAAAGUGCUGUCCUCAAUUAGAAUUAGAUAAGGAGAAG__AAAAAUGCGCCAUAAAGCCCAAGGAUUAAGCCAGAUUGAACAUCUUGGCGAGUCUGUGUGGCCCAGAGACAAUUUGCAUA ....(((((((((((..(((........))).))).....................(((((.(((((..(((((((.....))).)))).))))).)).))).....))))....)))). (-22.14 = -22.14 + 0.00)

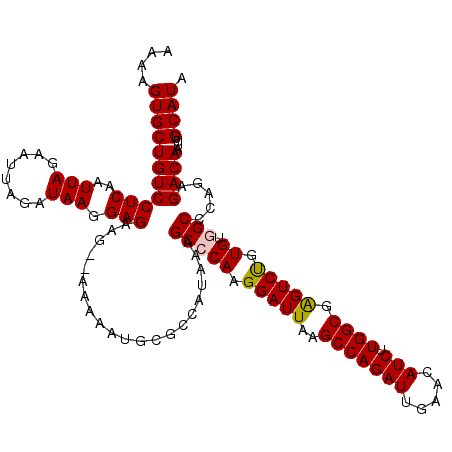
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:28 2006