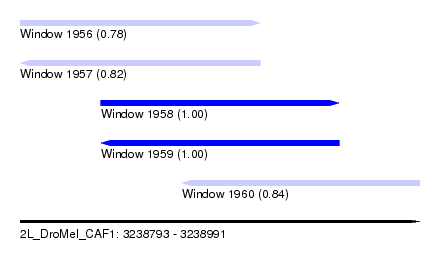
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,238,793 – 3,238,991 |
| Length | 198 |
| Max. P | 0.999974 |

| Location | 3,238,793 – 3,238,912 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -28.32 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238793 119 + 22407834 AACAUAGCUGUCGAAUGAAUCGCAAAUCGAGUGAACACAAUACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUU-UCAAAUCGGAUUCAGCGAGU ((((..((((.(((((((((.(((..(((......(((.......))).)))..))).))).))))))))))..))))..((((((((..((.......-.......))..)))))))). ( -33.34) >DroSec_CAF1 72989 120 + 1 AACACAGCUGUCGAAUGAAUCGCAAAUCGAGUGAACACAAUACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUUUUCAAAUCGGAUUCAGCGAGU ((((..((((.(((((((((.(((..(((......(((.......))).)))..))).))).))))))))))..))))..((((((((..((...............))..)))))))). ( -32.46) >DroSim_CAF1 74680 119 + 1 AACACAGCUGUCGAAUGAAUCGCAAAUCGAGUGAACACAAUACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUU-UCAAAUCGGAUUCAGCGAGU ((((..((((.(((((((((.(((..(((......(((.......))).)))..))).))).))))))))))..))))..((((((((..((.......-.......))..)))))))). ( -32.54) >DroEre_CAF1 69934 120 + 1 UACACAGCUGUCGAAUGGCUCGUAAAUCGAGUGAACACAAUACGAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCUCAACAUUUUUCAAAUCGAAUUCAGCGAGU ......((((((((.(.(((((.....))))).).........(((((((((((.((((((((....)))).).)))..)))))))..)))).............))))...)))).... ( -32.60) >DroYak_CAF1 68962 120 + 1 CACACAGCUGUCGAAUGCCUCGCAAAUCGAGGGAACACAAUACAAGUGUUGCAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGUCCAACAAUUUUCAAAUCGGAUUUAGCGAGU .(((..((((.(((((((((((.....))))(.(((((.......))))).).........)))))))))))..)))...((((((((((((...............)))))))))))). ( -39.36) >consensus AACACAGCUGUCGAAUGAAUCGCAAAUCGAGUGAACACAAUACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUUUUCAAAUCGGAUUCAGCGAGU .(((..((((.((((((...((((..(((......(((.......))).)))..))).)...))))))))))..)))...(((((((((.((...............)).))))))))). (-28.32 = -27.72 + -0.60)


| Location | 3,238,793 – 3,238,912 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -28.00 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238793 119 - 22407834 ACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUAUUGUGUUCACUCGAUUUGCGAUUCAUUCGACAGCUAUGUU ..(((((((..(((((....-....)))))..)))))))...((((((.(((..(((((((.((((..........)))))))))))...((((............))))))))).)))) ( -32.00) >DroSec_CAF1 72989 120 - 1 ACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUAUUGUGUUCACUCGAUUUGCGAUUCAUUCGACAGCUGUGUU ..(((((((..(((((.(....)..)))))..)))))))......((.((.((.(((((((.((((..........))))))))))).........(((((.....))).)))).)).)) ( -31.50) >DroSim_CAF1 74680 119 - 1 ACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUAUUGUGUUCACUCGAUUUGCGAUUCAUUCGACAGCUGUGUU ..(((((((..(((((....-....)))))..)))))))......((.((.((.(((((((.((((..........))))))))))).........(((((.....))).)))).)).)) ( -32.10) >DroEre_CAF1 69934 120 - 1 ACUCGCUGAAUUCGAUUUGAAAAAUGUUGAGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUCGUAUUGUGUUCACUCGAUUUACGAGCCAUUCGACAGCUGUGUA ..(((((((.((((((((....)).)))))).)))))))................(((((.(((..........((((((...((......))...))))))........))).))))). ( -29.57) >DroYak_CAF1 68962 120 - 1 ACUCGCUAAAUCCGAUUUGAAAAUUGUUGGACUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUGCAACACUUGUAUUGUGUUCCCUCGAUUUGCGAGGCAUUCGACAGCUGUGUG ..(((((.(.((((((.(....)..)))))).).)))))................(((((.((((..((((((((.......)))))..((((.....)))))))...).))).))))). ( -31.00) >consensus ACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUAUUGUGUUCACUCGAUUUGCGAUUCAUUCGACAGCUGUGUU ..(((((((.((((((.........)))))).)))))))....(((((.(((..(((((((.((((..........)))))))))))...((((............))))))))).))). (-28.00 = -27.92 + -0.08)


| Location | 3,238,833 – 3,238,951 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -36.61 |
| Consensus MFE | -34.26 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238833 118 + 22407834 UACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUU-UCAAAUCGGAUUCAGCGAGUUUUUUCGCACUUGCUAAGUA-UUGCAGGUCCUCUGCUUGG ..((((..(.(((.((((((.(((..(((............(((((((..((.......-.......))..)))))))(......))))..))).....)-)))))..))).)..)))). ( -38.54) >DroSec_CAF1 73029 120 + 1 UACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUUUUCAAAUCGGAUUCAGCGAGUUUUUUCGCACUUGCUAAGUUUUUGCAGAUACUCCGCUUGG ..(((((((.(...(((((..((....((((((.......((((((((..((...............))..)))))))).......).)).)))...))..)))))....).))))))). ( -36.50) >DroSim_CAF1 74720 118 + 1 UACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUU-UCAAAUCGGAUUCAGCGAGUUUUUUCGCACUUGCUAAGUUUUUGCAGGUCCU-CGCUUGG ..(((((((((((((((((......)))))..........((((((((..((.......-.......))..))))))))...))))))((((((.........))))))...-)))))). ( -38.14) >DroEre_CAF1 69974 120 + 1 UACGAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCUCAACAUUUUUCAAAUCGAAUUCAGCGAGUUUUUUCGCACUUGCUAAGUUUUUGCGGGUUCUCCACUUGG ..(((((((.(((.(((((..((....((((((.......(((((((((.((...............)).))))))))).......).)).)))...))..)))))..))).))))))). ( -36.40) >DroYak_CAF1 69002 120 + 1 UACAAGUGUUGCAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGUCCAACAAUUUUCAAAUCGGAUUUAGCGAGUUUUUUCGCACUUGCUAAGUUUUUGCGGGUUCUCUACGUGG ..(((((((((((((((....))).)))))..........((((((((((((...............)))))))))))).......)))))))........(..((.........))..) ( -33.46) >consensus UACAAGUGGCGGAAUGCAGUUGCGUUUGCAGUCUUGUUUUUUCGCUGAGCCCAACAAUUUUCAAAUCGGAUUCAGCGAGUUUUUUCGCACUUGCUAAGUUUUUGCAGGUCCUCCGCUUGG ..(((((((.(((.(((((..((....((((((.......(((((((((.((...............)).))))))))).......).)).)))...))..)))))..))).))))))). (-34.26 = -33.82 + -0.44)



| Location | 3,238,833 – 3,238,951 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238833 118 - 22407834 CCAAGCAGAGGACCUGCAA-UACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUA ....((((.....))))..-......(((((((((.......(((((((..(((((....-....)))))..)))))))........((.((((........)))).)))).))))))). ( -39.40) >DroSec_CAF1 73029 120 - 1 CCAAGCGGAGUAUCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUA ....((((.....)))).........(((((((((.......(((((((..(((((.(....)..)))))..)))))))........((.((((........)))).)))).))))))). ( -38.70) >DroSim_CAF1 74720 118 - 1 CCAAGCG-AGGACCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUA ....(((-......))).........(((((((((.......(((((((..(((((....-....)))))..)))))))........((.((((........)))).)))).))))))). ( -35.80) >DroEre_CAF1 69974 120 - 1 CCAAGUGGAGAACCCGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUUCGAUUUGAAAAAUGUUGAGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUCGUA ....((((.....)))).........((.((((((.......(((((((.((((((((....)).)))))).)))))))........((.((((........)))).)))).)))).)). ( -31.90) >DroYak_CAF1 69002 120 - 1 CCACGUAGAGAACCCGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUAAAUCCGAUUUGAAAAUUGUUGGACUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUGCAACACUUGUA ...(((.(((.............(((((.(((........))).))))).((((((.(....)..))))))))).))).....(((((..(((((..((....)).)))))...))))). ( -28.60) >consensus CCAAGCGGAGAACCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAAAAAACAAGACUGCAAACGCAACUGCAUUCCGCCACUUGUA ....((((.....)))).........(((((((((.......(((((((.((((((.........)))))).)))))))........((.((((........)))).)))).))))))). (-31.76 = -32.00 + 0.24)


| Location | 3,238,873 – 3,238,991 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.07 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.64 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238873 118 - 22407834 AUCGAACAUAAAAAAGGUGGAAGCCGUUAAACGCCUAAAUCCAAGCAGAGGACCUGCAA-UACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAA .(((.((.......(((((............)))))........((((.....))))..-..........)).)))......(((((((..(((((....-....)))))..))))))). ( -33.30) >DroSec_CAF1 73069 120 - 1 AUCGAACAUAAAAAAGAUGGAAGCCCUUAAAUGCCUAAAUCCAAGCGGAGUAUCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAA .(((.((..........((((.((........)).....)))).((((.....)))).............)).)))......(((((((..(((((.(....)..)))))..))))))). ( -29.40) >DroSim_CAF1 74760 102 - 1 AUCG----------------AAGCCGUUAAAUGCCUAAAUCCAAGCG-AGGACCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGA-AAUUGUUGGGCUCAGCGAA .(((----------------.....(((((.(((.....(((.....-.)))...))).....))))).....)))......(((((((..(((((....-....)))))..))))))). ( -25.80) >DroEre_CAF1 70014 120 - 1 AUUGAACAUAAAAAUGCUGGAAGCCGUUAAAUGCCUAAAUCCAAGUGGAGAACCCGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUUCGAUUUGAAAAAUGUUGAGCUCAGCGAA ..............(((((((((.(((...))).))...)))..((((.....))))........)))).............(((((((.((((((((....)).)))))).))))))). ( -26.10) >DroYak_CAF1 69042 120 - 1 AUCGAACAUAAAAAAGAUGGAAACCUUUGAAUGCCUAAAUCCACGUAGAGAACCCGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUAAAUCCGAUUUGAAAAUUGUUGGACUCAGCGAA ......(((...((((..(....)))))..)))..........(((.(((.............(((((.(((........))).))))).((((((.(....)..))))))))).))).. ( -21.10) >consensus AUCGAACAUAAAAAAGAUGGAAGCCGUUAAAUGCCUAAAUCCAAGCGGAGAACCUGCAAAAACUUAGCAAGUGCGAAAAAACUCGCUGAAUCCGAUUUGAAAAUUGUUGGGCUCAGCGAA ...............................(((..........((((.....))))....(((.....)))))).......(((((((.((((((.........)))))).))))))). (-21.16 = -20.64 + -0.52)

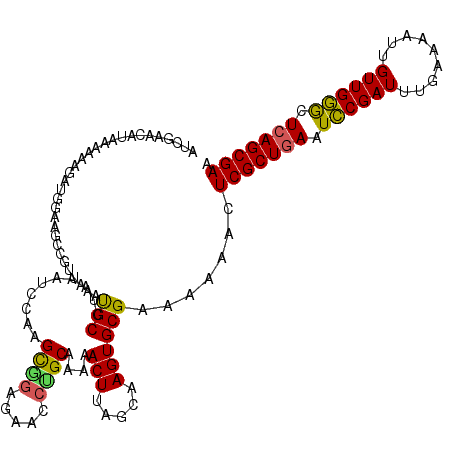
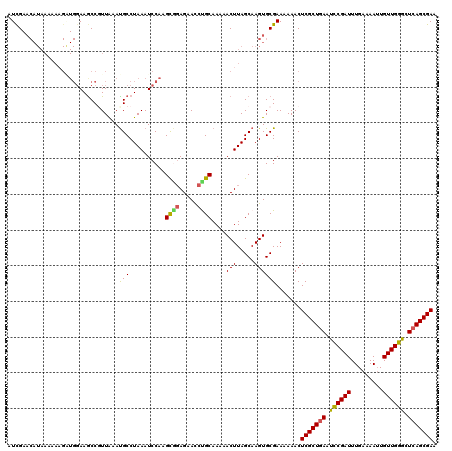
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:21 2006