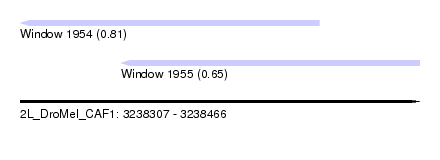
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,238,307 – 3,238,466 |
| Length | 159 |
| Max. P | 0.812181 |

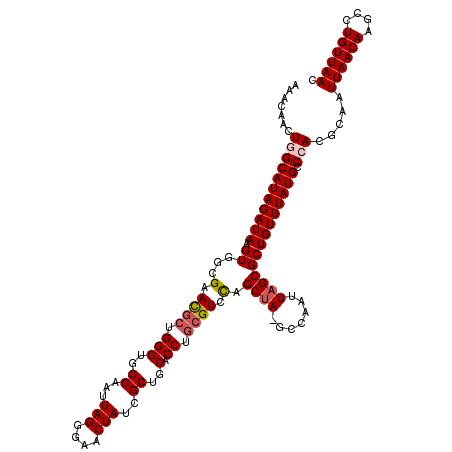
| Location | 3,238,307 – 3,238,426 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238307 119 - 22407834 AAACAACUGGCAUAGAUAAGUGGCGAACUCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUCUAGCUA-GCCAAUUAGCGCUGUUUAUGCCCACGCAAUUAACAAGCCUGUUAAC ........((((((((((.((((((......(((..((...(((....)))..))..).))..))))..((((-(....))))))))))))))))).......((((((....)))))). ( -34.50) >DroSec_CAF1 72483 119 - 1 AAACAACUAGCAUAGAUAAGUGCCGAACGAUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUACAGCUA-GCCAAUUAGCGCUGUUUAUGCCCACGCAAUUAACAAGCCUGUUAAC .........(((((((((.((((..(....(((.((((...(((....))).(((.......)))....))))-.))).)..)))))))))))))........((((((....)))))). ( -32.40) >DroSim_CAF1 74173 119 - 1 AAACAACUAGCAUAGAUAAGUGGCGAACGCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUACAGCUA-GCCAAUUAGCGCUGUUUAUGCCCACGCAAUUAACAAGCCUGUUAAC .........((((((((....((((.((((.(((..((...(((....)))..))..).)).)))).).((((-(....))))))))))))))))........((((((....)))))). ( -35.40) >DroEre_CAF1 69451 120 - 1 CAACAACUGGCAUAGAUAAGUGGCGAAUGCUGGGUGGCAAUUAGGAAACUAUCGCUGCACCUGCGUCCAGCAAUCCCAAUUAGCGCUGUUUAUGGCCACGCAAUUAACAAGCCUGUUAAC ....(((.(((.....(((.(((.((.(((((((.(((...(((....)))..)))((....)).))))))).))))).)))(((..((.....))..))).........))).)))... ( -36.20) >DroYak_CAF1 68446 120 - 1 UAACAACUGGCAUAGAUAAGUGCCGAAUGCUGGGUGGCAAUUAGGAAACUAUCGCUGCACCUGCGUCCAGCUAUGCCAAUUAGCGCUGUUUAUGGCCACGCAAUUAACAAGCCUGUUAAC (((((..((((((......))))))...(((((.(((((..(((....)))..((((.((....)).))))..))))).)))))((((((....((...))....))).))).))))).. ( -36.70) >consensus AAACAACUGGCAUAGAUAAGUGGCGAACGCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUCCAGCUA_GCCAAUUAGCGCUGUUUAUGCCCACGCAAUUAACAAGCCUGUUAAC .......(((((((((((.((...(.((((.(((..((...(((....)))..))..).)).)))).).((((.......)))))))))))))).))).....((((((....)))))). (-28.98 = -29.58 + 0.60)

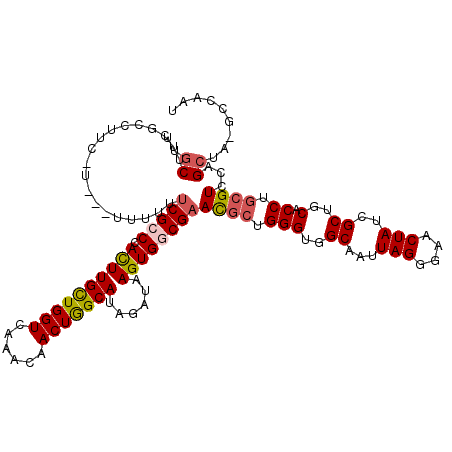
| Location | 3,238,347 – 3,238,466 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3238347 119 - 22407834 UUGCUUCGCCUUCCUUUUUUUUUUCGCCCACUUGCUGGUCAAACAACUGGCAUAGAUAAGUGGCGAACUCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUCUAGCUA-GCCAAU ..(((.(((.............((((((.((((((((((((......)))).))).)))))))))))....(((..((...(((....)))..))..).)).)))...)))..-...... ( -36.30) >DroSec_CAF1 72523 114 - 1 UUGCUUCGACUUC-----UUUUUUCGCCCACUUGCUGGUCAAACAACUAGCAUAGAUAAGUGCCGAACGAUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUACAGCUA-GCCAAU .............-----.......((((((((((((((......))))))).))....((.....))..)))))(((...(((....)))..((((.((....)).))))..-)))... ( -29.00) >DroSim_CAF1 74213 114 - 1 UUGCUUCGACUUC-----UUUUUUCGCCCACUUGCUGGUCAAACAACUAGCAUAGAUAAGUGGCGAACGCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUACAGCUA-GCCAAU ..(((........-----.....(((((.((((((((((......)))))))......))))))))((((.(((..((...(((....)))..))..).)).))))..)))..-...... ( -37.00) >DroEre_CAF1 69491 113 - 1 ---CUUUUCCUUCUU----UUUUUCGCCCAUUUGUUGGUCCAACAACUGGCAUAGAUAAGUGGCGAAUGCUGGGUGGCAAUUAGGAAACUAUCGCUGCACCUGCGUCCAGCAAUCCCAAU ---............----...((((((.(((((((.(.(((.....))))...)))))))))))))(((((((.(((...(((....)))..)))((....)).)))))))........ ( -35.20) >DroYak_CAF1 68486 117 - 1 UUGCCUCGCCUUCAU---UUUUUCCGCCCAUUUGCUGGUCUAACAACUGGCAUAGAUAAGUGCCGAAUGCUGGGUGGCAAUUAGGAAACUAUCGCUGCACCUGCGUCCAGCUAUGCCAAU ..((((.((.(((..---.....((((......)).))..........(((((......)))))))).)).))))((((..(((....)))..((((.((....)).))))..))))... ( -34.10) >consensus UUGCUUCGCCUUC_U___UUUUUUCGCCCACUUGCUGGUCAAACAACUGGCAUAGAUAAGUGGCGAACGCUGGGUGGCAAUUAGGGAACUAUCGCUGCACCUGCGUCCAGCUA_GCCAAU ..((...................(((((.((((((((((......)))))))......))))))))((((.(((..((...(((....)))..))..).)).))))...))......... (-26.96 = -27.28 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:16 2006