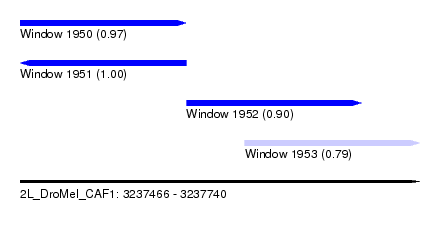
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,237,466 – 3,237,740 |
| Length | 274 |
| Max. P | 0.998572 |

| Location | 3,237,466 – 3,237,580 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -8.44 |
| Energy contribution | -8.80 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3237466 114 + 22407834 UGCAAUCGAAAGAUAAGCACAUUUAAAUGGAACUUUUUCCAGCUU------CUUUCUGGCUUAAGACUGAUGCUGUUAGCCUGAUUCGCUCCAUUUAACCUUAAUAUUCACUUUGCUUCA .((((((....)))........(((((((((........((((.(------(..(((......)))..)).))))...((.......)))))))))))...............))).... ( -24.30) >DroSec_CAF1 71597 113 + 1 UGCAAUC-AAAGUUAAACACAUUUAAAUGGAACUUUUUCCAGAUG------CUUUCUGGCUUAAGACUGAGUCUGUUAGCCAGAUUCGCUCCACUUAACCUUAAUAUUCACUUUCCUUCA .......-...(((((...........(((((....)))))((.(------(..(((((((..((((...))))...)))))))...))))...)))))..................... ( -23.30) >DroSim_CAF1 73290 113 + 1 UGCAAUC-AAAGUUAAACACAUUUAAAUGGAACUUUUUCCAGAUG------CUUUCUGGCUUAAGACUGAGUCUGUAAGCCAGAUUCGCUCCACUUAACCUUAAUAUUCACUUUCCUUCA .......-...(((((...........(((((....)))))((.(------(..(((((((((((((...)))).)))))))))...))))...)))))..................... ( -24.50) >DroEre_CAF1 68582 102 + 1 ------------------CCAUUUAAGUGGAACUUGUUUCAACCUCUCUACCUAUCUGGCUUAAGACUGAGGCUGUUAGCCCGAUUCGCUCCACUUAACCUUAAUAUUCACUUUCCUUCA ------------------....(((((((((...((..((....(((..(((.....)).)..)))..(.(((.....))))))..)).)))))))))...................... ( -17.70) >DroYak_CAF1 67585 116 + 1 UGCCAUA-CCAUUGCCAACCAUUUAAAUGGAACUUUACUCAACCAUUCAACCUAUCUGGCUUAAGACUGAGGCUGUUAGCCUCAUUCACCCCACUUAACCUU---GUUCACUUUCCUCCA .......-.....((((........(((((............))))).........))))....((.((((((.....)))))).))...............---............... ( -18.83) >consensus UGCAAUC_AAAGUUAAACACAUUUAAAUGGAACUUUUUCCAGCUG______CUUUCUGGCUUAAGACUGAGGCUGUUAGCCAGAUUCGCUCCACUUAACCUUAAUAUUCACUUUCCUUCA ...........................(((((....))))).............((.((((..((.......))...)))).)).................................... ( -8.44 = -8.80 + 0.36)


| Location | 3,237,466 – 3,237,580 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.86 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3237466 114 - 22407834 UGAAGCAAAGUGAAUAUUAAGGUUAAAUGGAGCGAAUCAGGCUAACAGCAUCAGUCUUAAGCCAGAAAG------AAGCUGGAAAAAGUUCCAUUUAAAUGUGCUUAUCUUUCGAUUGCA ....((((..((((...((((.(((((((((((((.....((.....)).....)).....((((....------...)))).....))))))))))).....))))...)))).)))). ( -28.50) >DroSec_CAF1 71597 113 - 1 UGAAGGAAAGUGAAUAUUAAGGUUAAGUGGAGCGAAUCUGGCUAACAGACUCAGUCUUAAGCCAGAAAG------CAUCUGGAAAAAGUUCCAUUUAAAUGUGUUUAACUUU-GAUUGCA ......(((((((((((.....(((((((((((((.((((.....)))).)).........(((((...------..))))).....)))))))))))..)))))).)))))-....... ( -31.90) >DroSim_CAF1 73290 113 - 1 UGAAGGAAAGUGAAUAUUAAGGUUAAGUGGAGCGAAUCUGGCUUACAGACUCAGUCUUAAGCCAGAAAG------CAUCUGGAAAAAGUUCCAUUUAAAUGUGUUUAACUUU-GAUUGCA ......(((((((((((.....(((((((((((...(((((((((.((((...)))))))))))))...------(....)......)))))))))))..)))))).)))))-....... ( -33.90) >DroEre_CAF1 68582 102 - 1 UGAAGGAAAGUGAAUAUUAAGGUUAAGUGGAGCGAAUCGGGCUAACAGCCUCAGUCUUAAGCCAGAUAGGUAGAGAGGUUGAAACAAGUUCCACUUAAAUGG------------------ ......................(((((((((((......(((.....)))(((..(((..(((.....)))...)))..))).....)))))))))))....------------------ ( -28.50) >DroYak_CAF1 67585 116 - 1 UGGAGGAAAGUGAAC---AAGGUUAAGUGGGGUGAAUGAGGCUAACAGCCUCAGUCUUAAGCCAGAUAGGUUGAAUGGUUGAGUAAAGUUCCAUUUAAAUGGUUGGCAAUGG-UAUGGCA .........((.(((---....((((((((..(((.((((((.....)))))).))...(((((...........))))).......)..))))))))...))).)).....-....... ( -31.00) >consensus UGAAGGAAAGUGAAUAUUAAGGUUAAGUGGAGCGAAUCAGGCUAACAGCCUCAGUCUUAAGCCAGAAAG______AAGCUGGAAAAAGUUCCAUUUAAAUGUGUUUAACUUU_GAUUGCA ......................(((((((((((......((((.........)))).....((((.............)))).....)))))))))))...................... (-15.34 = -14.86 + -0.48)

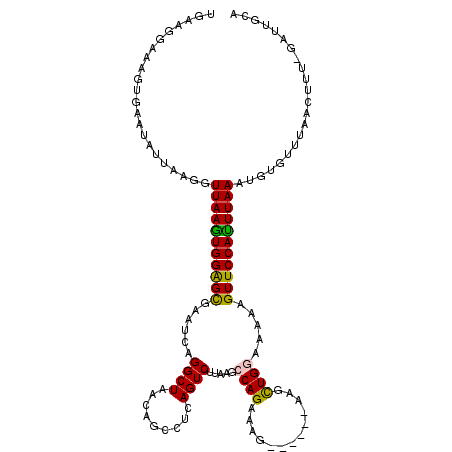
| Location | 3,237,580 – 3,237,700 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -35.78 |
| Energy contribution | -36.26 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3237580 120 + 22407834 AUCCUCUACAGUUCUGCUGCUUAUCUUUAGUUGCAGACAGCAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAUGUCGACUGGCCUGUCAGCCUCCGGAGCACUG ........(((((((((.(((.......))).))))).........((((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))))...)))) ( -39.10) >DroSec_CAF1 71710 120 + 1 AUCCUCUACAGUUCUGCUGCUUAUCUUUAGUUGCAGACAGGAUUAACCGGCACGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUG .........(((((((((((............)))).)))))))..((((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))))....... ( -40.40) >DroSim_CAF1 73403 120 + 1 AUCCUCUACAGUUCUGCUGCUUAUCUUUAGUUGCAGACAGCAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUG ........(((((((((.(((.......))).))))).........((((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))))...)))) ( -39.10) >DroEre_CAF1 68684 120 + 1 AUCCUCUCCAGCUCUGCUGCUUAUCUUUAGCUGCAGACAGCACCAAGCGGCGUGGCAGGAAGUCAUCCUGCUGCCGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCCCCGAAGCACUG ..........(((.(((.(((.......))).)))(((((.((...(((((..(((((((.....))))))))))))..)))))))....(((...(((......)))..)))))).... ( -40.40) >DroYak_CAF1 67701 120 + 1 AUCCUCUCCAGUUCUGCUGCUUAUCUUCAGUUGCAGACAGCAUUAACCGGCUUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCCCCGAAGCACUG ........(((((((((.(((.......))).)))))............(((((((((((.....)))))))((((.(((((.(((......))).)).))).)))).....)))))))) ( -35.80) >consensus AUCCUCUACAGUUCUGCUGCUUAUCUUUAGUUGCAGACAGCAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUG ........(((((((((.(((.......))).))))).........((((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))))...)))) (-35.78 = -36.26 + 0.48)



| Location | 3,237,620 – 3,237,740 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -38.42 |
| Energy contribution | -38.14 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3237620 120 + 22407834 CAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAUGUCGACUGGCCUGUCAGCCUCCGGAGCACUGUCCAUUUUUUAGGGCACCUCGACGAGGGCACAGGUGUUCC ........((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))((((((((((((........)))).((((...)))).....)))))))) ( -43.20) >DroSec_CAF1 71750 120 + 1 GAUUAACCGGCACGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUGUGCAUUUUUUAGGGCACCUCGACGAGGGCACAGGUGUUCC ........(((.((((((((.....))))))))..(((((((..........)))))))......)))...((((((((((((.(((.(..((....))..).))).))))).))))))) ( -44.40) >DroSim_CAF1 73443 120 + 1 CAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUGUGCAUUUUUUAGGGCACCUCGACGAGGGCACAGGUGUUCC ........((...(((((((.....)))))))((((.(((((.(((......))).)).))).))))..))((((((((((((.(((.(..((....))..).))).))))).))))))) ( -43.50) >DroEre_CAF1 68724 120 + 1 CACCAAGCGGCGUGGCAGGAAGUCAUCCUGCUGCCGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCCCCGAAGCACUGUGCAUUUUUUAGGGCACCUCGACGGGGGCACAGGUGUUCC ......(((((..(((((((.....))))))))))))(((((..........)))))((((((..(((((((..(....((((..........))))..)..)))))))))))))..... ( -51.70) >DroYak_CAF1 67741 117 + 1 CAUUAACCGGCUUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCCCCGAAGCACUGUGCAUUUUUU---GCACCUCGCCGAGGGCACAGGUGUUCC .......((((...((((((.....)))))).)))).(((((..........)))))((((((..((((.((..((...(((((.....)---))))...)))).))))))))))..... ( -42.80) >consensus CAUUAACCGGCAUGGCAGGAAGUCAUCCUGUUGCUGUCAGUCUGUCAAAGUCGACUGGCCUGUCAGCCUCCGGAGCACUGUGCAUUUUUUAGGGCACCUCGACGAGGGCACAGGUGUUCC .......(((((..((((((.....))))))))))).(((((..........)))))((((((..((((.((..(....((((..........))))..)..)).))))))))))..... (-38.42 = -38.14 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:14 2006