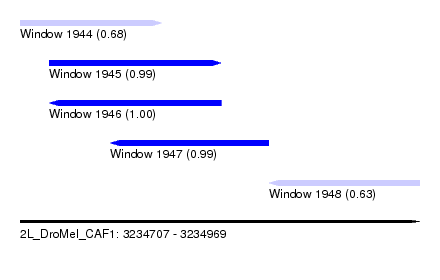
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,234,707 – 3,234,969 |
| Length | 262 |
| Max. P | 0.996422 |

| Location | 3,234,707 – 3,234,800 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.33 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234707 93 + 22407834 -------------GCCACAGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAACGCCGGCGAA -------------((....)).................(((...(((.....)))..(((((.(......((((-...----(....)...))))......).)))))))) ( -19.50) >DroSec_CAF1 68800 93 + 1 -------------GCCACUGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAACGCCGGCGAA -------------((....)).................(((...(((.....)))..(((((.(......((((-...----(....)...))))......).)))))))) ( -19.30) >DroSim_CAF1 70525 93 + 1 -------------GCCACUGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAGCGCCGGCGAA -------------((....)).................(((...(((.....)))..(((((.((.....((((-...----(....)...)))).....)).)))))))) ( -22.90) >DroEre_CAF1 65826 106 + 1 UCCGCCUCGUGUGGCAGUGGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAG----CGAAAGAAAGAAAUUGAAGCGCCGGCGAA ..((((..(((((((((((....))).)).))..(((((((...(((.....)))..((((..(......)..)-).)----)........)))))))..)))).)))).. ( -34.60) >DroYak_CAF1 64686 106 + 1 -----GUGCCGUUGCAGCGGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUCCGAACGAAAGAAAGAAAGAAAUUGAAGCGCCGGCGAA -----.((((((....))))))................(((...(((.....)))..(((((.((..((((((.(...(....)...)...))))))...)).)))))))) ( -29.90) >consensus _____________GCCACGGCAACACAUGACCAACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC_GAA____CGAAAGAAAGAAAUUGAAGCGCCGGCGAA .............((....)).................(((...(((.....)))..(((((.((..((((((..................))))))...)).)))))))) (-16.93 = -17.33 + 0.40)

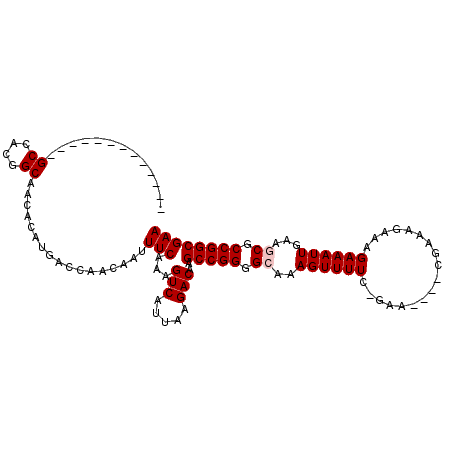

| Location | 3,234,726 – 3,234,839 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -29.01 |
| Energy contribution | -29.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234726 113 + 22407834 AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAACGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGCUGCAGC- ..((((((....(((.....)))..(((((.(......((((-...----(....)...))))......).)))))))))))((.(((..((((.(((....))).)))))))))...- ( -30.30) >DroSec_CAF1 68819 113 + 1 AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAACGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGCUGCAGC- ..((((((....(((.....)))..(((((.(......((((-...----(....)...))))......).)))))))))))((.(((..((((.(((....))).)))))))))...- ( -30.30) >DroSim_CAF1 70544 113 + 1 AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAA----CGAAAGAAAGAAAUUGAAGCGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGCUGCAGC- ..((((((....(((.....)))..(((((.((.....((((-...----(....)...)))).....)).)))))))))))((.(((..((((.(((....))).)))))))))...- ( -33.90) >DroEre_CAF1 65858 113 + 1 AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC-GAG----CGAAAGAAAGAAAUUGAAGCGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGCUGCAGC- ..((((((....(((.....)))..(((((.((.....((((-...----(....)...)))).....)).)))))))))))((.(((..((((.(((....))).)))))))))...- ( -35.00) >DroYak_CAF1 64713 119 + 1 AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUCCGAACGAAAGAAAGAAAGAAAUUGAAGCGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGUUGCAGCA ..((((((....(((.....)))..(((((.((..((((((.(...(....)...)...))))))...)).)))))))))))........(((((((.(........)...))))))). ( -34.20) >consensus AACAAUUUCAAAGUCAUUAAGACAAGCCGGGGCAAAGUUUUC_GAA____CGAAAGAAAGAAAUUGAAGCGCCGGCGAAUUGGCCAGCGAGCUGCAAACUUUUUUACGGCGCUGCAGC_ ..(((((((...(((.....)))..((....))..........................)))))))...(((.(((......))).))).((((((................)))))). (-29.01 = -29.01 + -0.00)


| Location | 3,234,726 – 3,234,839 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -31.14 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234726 113 - 22407834 -GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGUUUCAAUUUCUUUCUUUCG----UUC-GAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU -((((((((...........).))))))).(((((((......)))))))...(((((((....((((.----...-)))).....((....))..(((.....)))...))))))).. ( -31.70) >DroSec_CAF1 68819 113 - 1 -GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGUUUCAAUUUCUUUCUUUCG----UUC-GAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU -((((((((...........).))))))).(((((((......)))))))...(((((((....((((.----...-)))).....((....))..(((.....)))...))))))).. ( -31.70) >DroSim_CAF1 70544 113 - 1 -GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG----UUC-GAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU -((((((((...........).))))))).(((((((......)))))))...(((((((....((((.----...-)))).....((....))..(((.....)))...))))))).. ( -31.50) >DroEre_CAF1 65858 113 - 1 -GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG----CUC-GAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU -((((((((...........).))))))).(((((((......)))))))...(((((((....((((.----...-)))).....((....))..(((.....)))...))))))).. ( -31.50) >DroYak_CAF1 64713 119 - 1 UGCUGCAACGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCUUUCGUUCGGAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU .((((((((...........).))))))).(((((((......)))))))...(((((((....(((((.......))))).....((....))..(((.....)))...))))))).. ( -31.40) >consensus _GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG____UUC_GAAAACUUUGCCCCGGCUUGUCUUAAUGACUUUGAAAUUGUU .((((((((...........).))))))).(((((((......)))))))...(((((((..........................((....))..(((.....)))...))))))).. (-31.14 = -30.98 + -0.16)



| Location | 3,234,766 – 3,234,870 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.40 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234766 104 - 22407834 UGAAUGAUGAUUUUGUGGCCAUUUUGAUGCU------GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGUUUCAAUUUCUUUCUUUCG----UUC-GA .((((((.((..(((..((......((.(((------((...(((...........))).)))))))((((((......))))))))..)))..))......)))----)))-.. ( -30.40) >DroSec_CAF1 68859 103 - 1 UGAAUGAUGAUUUUGUGGCCAUUUUGACG-U------GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGUUUCAAUUUCUUUCUUUCG----UUC-GA .((((((.((..(((..((.........(-.------((((((((...........).))))))).)((((((......))))))))..)))..))......)))----)))-.. ( -31.30) >DroSim_CAF1 70584 104 - 1 UGAAUGAUGAUUUUGUGGCCAUUUUGAUGCU------GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG----UUC-GA .((((((.((..(((.(((......((.(((------((...(((...........))).)))))))((((((......))))))))).)))..))......)))----)))-.. ( -32.40) >DroEre_CAF1 65898 104 - 1 UGAAUGAUGAUUUUGUGGCCAUUUUGGUGCU------GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG----CUC-GA .(((.((.((..(((.(((......((((((------(...)))))))(((((....))))).....((((((......))))))))).)))..))..)).))).----...-.. ( -33.50) >DroYak_CAF1 64753 115 - 1 UGAAUGAUGAUUUUGUGGCCAUUUUGAUGCUGCUGCUGCUGCAACGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCUUUCGUUCGGA .((((((.((..(((.(((......((.(((((.(((..(((......)))....)))..)))))))((((((......))))))))).)))..))..........))))))... ( -34.40) >consensus UGAAUGAUGAUUUUGUGGCCAUUUUGAUGCU______GCUGCAGCGCCGUAAAAAAGUUUGCAGCUCGCUGGCCAAUUCGCCGGCGCUUCAAUUUCUUUCUUUCG____UUC_GA .(((.((.((..(((..((..................((((((((...........).)))))))..((((((......))))))))..)))..))..)).)))........... (-29.80 = -29.40 + -0.40)

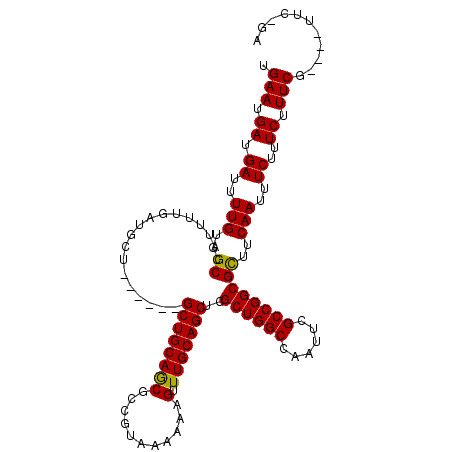
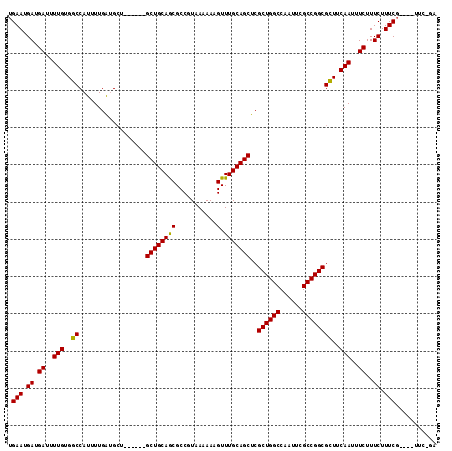
| Location | 3,234,870 – 3,234,969 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -25.15 |
| Energy contribution | -26.52 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234870 99 - 22407834 CGACGCAUGACGAGCUGCAGCGCCAGUUGCAUUCGAGUUUCAAUUACUCGAAUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCGGA--------------------- ...........(((((((..(.((((..((((((((((.......))))))))))...)))).).((((.((.....)).))))...).))))))....--------------------- ( -35.40) >DroSec_CAF1 68962 99 - 1 CGACGCAUGACGAGCUGCAGCGCCAGUUGCAUUCGAGUUUCAAUUACUCGAAUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCGAG--------------------- ...........(((((((..(.((((..((((((((((.......))))))))))...)))).).((((.((.....)).))))...).))))))....--------------------- ( -35.40) >DroSim_CAF1 70688 99 - 1 CGACGCAUGACGAGCUGCAGCGCCAGUUGCAUUCGAGUUUCAAUUACUCGAAUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCGAG--------------------- ...........(((((((..(.((((..((((((((((.......))))))))))...)))).).((((.((.....)).))))...).))))))....--------------------- ( -35.40) >DroEre_CAF1 66002 99 - 1 UGACGCAUGACGAGCUGCAGCGCCAGCUGCAUUCCAGUUUCAAUUACUCGAAUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCGAG--------------------- ...........(((((((..(.((((..((((((.(((.......))).))))))...)))).).((((.((.....)).))))...).))))))....--------------------- ( -29.50) >DroYak_CAF1 64868 99 - 1 UGACGCAUGACGAGCUGCAGCGCCAGUUGCAUUCCAGUUUCAAUUACUCGAUUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCUAG--------------------- ...........(((((((..(.((((..(((.((.(((.......))).)).)))...)))).).((((.((.....)).))))...).))))))....--------------------- ( -26.30) >DroPer_CAF1 80477 114 - 1 AGACGCAUGACGAGUGG------AAGUUGCAUUCGCAUUUCAAUUACUUCAACGACACAUGGGGCAAAGGGACCAAAUCGUGUUAAAGUCCGCUCCUGCAUCUCCACCAUCUCCAUCGUA ..(((.(((..((((((------(.(.((((.(((.................))).....(((((....((((..............))))))))))))).))))))..))..)))))). ( -24.47) >consensus CGACGCAUGACGAGCUGCAGCGCCAGUUGCAUUCGAGUUUCAAUUACUCGAAUGCCACCUGGGGAAAUGGGACAAAAUCGCAUUAAAGUCAGCUCCGAG_____________________ ...........((((((...(.((((.(((((((((((.......))))))))).)).)))).).((((.((.....)).)))).....))))))......................... (-25.15 = -26.52 + 1.36)

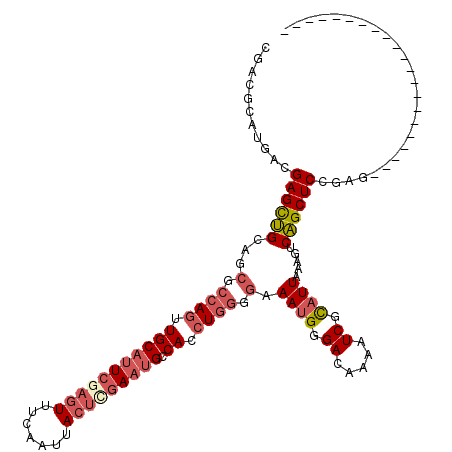

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:09 2006