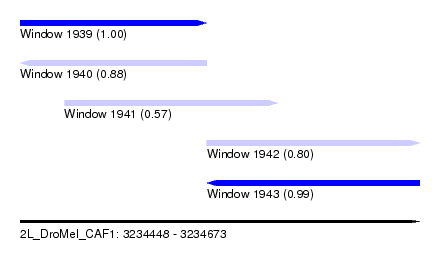
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,234,448 – 3,234,673 |
| Length | 225 |
| Max. P | 0.999541 |

| Location | 3,234,448 – 3,234,553 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -27.82 |
| Energy contribution | -29.54 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
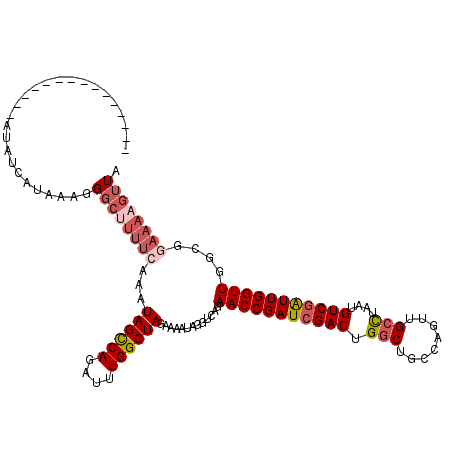
>2L_DroMel_CAF1 3234448 105 + 22407834 ---------------AUAUCAUAAAGGGCUUUUCAAAUAGUCAGAUUUGGCUAGAAAAUUGGGCAGAACCGAUCGACUGGCUGGCAGCUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUA ---------------...........((((((((...(((((......))))).........((..(((((((((((.....(((....)))....))))))))))).)).)))))))). ( -36.80) >DroSec_CAF1 68544 105 + 1 ---------------AUAGUAUAAAGGGCUUUUCAAAUAGCCAGAUCUGGCUAGCAAAUAUGUCAGAACCGAUCGACUGGCUGGCAGUUGCCUUAUGUCGAUUGGUUGGCGGAAAAGUUA ---------------...........((((((((...((((((....)))))).......(((((..((((((((((.....(((....)))....))))))))))))))))))))))). ( -40.70) >DroSim_CAF1 70269 105 + 1 ---------------AUUGCAUAAAGGGCUUUUCAUAUAGCCAGAUUUGGCUAGCAAAUAUGUCAGAACCGAUCGACUGGCUGACAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUA ---------------...........((((((((...(((((......))))).......(((((..((((((((((.(((........)))....))))))))))))))))))))))). ( -39.20) >DroEre_CAF1 65574 95 + 1 ---------------AUAUCGCA---G-------AUAUAGCCAGUUUUGGCUAGAAAACAGGGCAGAACCGAUCGACUGGCUGCCAGUGGGUUAAUGUCAGUUGGUUGGCGGAAAAGUUA ---------------...((((.---.-------.....(((.(((((......)))))..))).....((((((((((((.(((....)))....))))))))))))))))........ ( -31.50) >DroYak_CAF1 64416 117 + 1 GCCGUAAUUAAAACCAUUUCGCA---GGCAUUUAAAAUAGCCACAUUUGGCUAGAAAAUUGUUCAGAACCGAUCGACUGGCUGCCAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUA ...........(((..((((((.---((((.......((((((....))))))......))))...(((((((((((.(((........)))....))))))))))).))))))..))). ( -38.62) >consensus _______________AUAUCAUAAAGGGCUUUUCAAAUAGCCAGAUUUGGCUAGAAAAUAGGUCAGAACCGAUCGACUGGCUGCCAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUA ..........................((((((((...((((((....)))))).............(((((((((((.(((........)))....)))))))))))....)))))))). (-27.82 = -29.54 + 1.72)

| Location | 3,234,448 – 3,234,553 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234448 105 - 22407834 UAACUUUUCCGCCAACCAAUCGACAUUAGGCAGCUGCCAGCCAGUCGAUCGGUUCUGCCCAAUUUUCUAGCCAAAUCUGACUAUUUGAAAAGCCCUUUAUGAUAU--------------- ...((((((.((.((((.((((((....(((....))).....)))))).))))..)).........(((.((....)).)))...)))))).............--------------- ( -21.80) >DroSec_CAF1 68544 105 - 1 UAACUUUUCCGCCAACCAAUCGACAUAAGGCAACUGCCAGCCAGUCGAUCGGUUCUGACAUAUUUGCUAGCCAGAUCUGGCUAUUUGAAAAGCCCUUUAUACUAU--------------- ...((((((....((((.((((((....(((....))).....)))))).)))).............((((((....))))))...)))))).............--------------- ( -28.00) >DroSim_CAF1 70269 105 - 1 UAACUUUUCCGCCAACCAAUCGACAUUAGGCAACUGUCAGCCAGUCGAUCGGUUCUGACAUAUUUGCUAGCCAAAUCUGGCUAUAUGAAAAGCCCUUUAUGCAAU--------------- ...((((((....((((.((((((....(((........))).)))))).)))).............((((((....))))))...)))))).............--------------- ( -26.60) >DroEre_CAF1 65574 95 - 1 UAACUUUUCCGCCAACCAACUGACAUUAACCCACUGGCAGCCAGUCGAUCGGUUCUGCCCUGUUUUCUAGCCAAAACUGGCUAUAU-------C---UGCGAUAU--------------- .........(((.......................(((((((........))..)))))........((((((....))))))...-------.---.)))....--------------- ( -18.80) >DroYak_CAF1 64416 117 - 1 UAACUUUUCCGCCAACCAAUCGACAUUAGGCAACUGGCAGCCAGUCGAUCGGUUCUGAACAAUUUUCUAGCCAAAUGUGGCUAUUUUAAAUGCC---UGCGAAAUGGUUUUAAUUACGGC ..........(((((((.((((((....(((........))).)))))).)))).............((((((....))))))..(((((.(((---........))))))))....))) ( -29.40) >consensus UAACUUUUCCGCCAACCAAUCGACAUUAGGCAACUGCCAGCCAGUCGAUCGGUUCUGACCUAUUUUCUAGCCAAAUCUGGCUAUUUGAAAAGCCCUUUAUGAUAU_______________ .............((((.((((((....(((........))).)))))).)))).............((((((....))))))..................................... (-17.90 = -18.54 + 0.64)


| Location | 3,234,473 – 3,234,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -37.34 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234473 120 + 22407834 UCAGAUUUGGCUAGAAAAUUGGGCAGAACCGAUCGACUGGCUGGCAGCUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUAAAGAAAACUCGGUUGCAAUCAAGAGCAUUGAAGGGCGGCC ......(..(((((...(((((......)))))...)))))..)..((((((((((((...(((((((.(((...((((......))))...))))))))))..)))))...))))))). ( -38.60) >DroSec_CAF1 68569 120 + 1 CCAGAUCUGGCUAGCAAAUAUGUCAGAACCGAUCGACUGGCUGGCAGUUGCCUUAUGUCGAUUGGUUGGCGGAAAAGUUAAAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCC ((...((((((..........))))))((((((((((.....(((....)))....))))))))))....))...((((......)))).((((((..((((.....))))...)))))) ( -39.10) >DroSim_CAF1 70294 120 + 1 CCAGAUUUGGCUAGCAAAUAUGUCAGAACCGAUCGACUGGCUGACAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUAAAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCC ((..(((((.....)))))..((((..((((((((((.(((........)))....))))))))))))))))...((((......)))).((((((..((((.....))))...)))))) ( -37.70) >DroEre_CAF1 65589 120 + 1 CCAGUUUUGGCUAGAAAACAGGGCAGAACCGAUCGACUGGCUGCCAGUGGGUUAAUGUCAGUUGGUUGGCGGAAAAGUUAAAGAAAACUCUGUUGCAAUCAAGCGCAUUGAAGGACGGCC ((..(..(((((.((.(((((((.....(((((((((((((.(((....)))....)))))))))))))(......)..........)))))))....)).))).))..)..))...... ( -34.30) >DroYak_CAF1 64453 120 + 1 CCACAUUUGGCUAGAAAAUUGUUCAGAACCGAUCGACUGGCUGCCAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUAAAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCC .....(((.(((.(((.....)))..(((((((((((.(((........)))....)))))))))))))).))).((((......)))).((((((..((((.....))))...)))))) ( -37.00) >consensus CCAGAUUUGGCUAGAAAAUAGGUCAGAACCGAUCGACUGGCUGCCAGUUGCCUAAUGUCGAUUGGUUGGCGGAAAAGUUAAAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCC ........((((..............(((((((((((.(((........)))....))))))))))).((((...((((......))))...))))..((((.....)))).....)))) (-29.70 = -29.78 + 0.08)

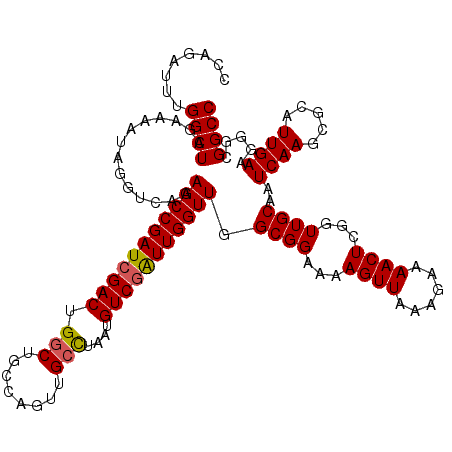
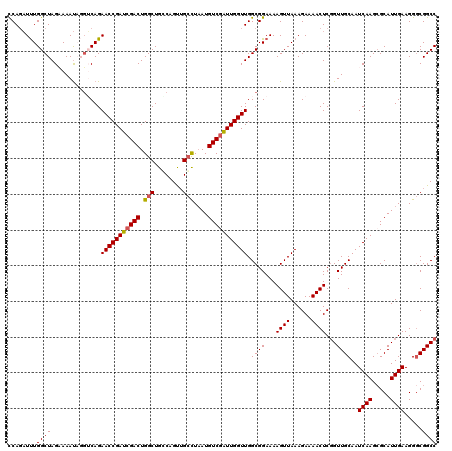
| Location | 3,234,553 – 3,234,673 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -35.52 |
| Energy contribution | -35.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234553 120 + 22407834 AAGAAAACUCGGUUGCAAUCAAGAGCAUUGAAGGGCGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAGUGGAUGCACUGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((((..(((((..((.((((.....(((.((((.((((.....((....)).))))(((((.....))))))))).)))..(.......))))))))))))..))))).... ( -36.40) >DroSec_CAF1 68649 117 + 1 AAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAG---CUGCACUGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((((..((((.((.((((.......(((.((((.((((.....((....)).))))((((..---..)))))))).)))..(.......))))).))))))..))))).... ( -37.70) >DroSim_CAF1 70374 117 + 1 AAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAG---CUGCACUGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((((..((((.((.((((.......(((.((((.((((.....((....)).))))((((..---..)))))))).)))..(.......))))).))))))..))))).... ( -37.70) >DroEre_CAF1 65669 117 + 1 AAGAAAACUCUGUUGCAAUCAAGCGCAUUGAAGGACGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAG---CUGCACUGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((..((((((.((.((((......((...((((.((((.....((....)).))))((((..---..))))))))..))..(.......))))).))))))))..))).... ( -32.60) >DroYak_CAF1 64533 117 + 1 AAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAA---CUGCACCGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((((..((((.((.((((.......(((.((((((.((.....)).)))......(((((..---..)))))))).)))..(.......))))).))))))..))))).... ( -39.40) >consensus AAGAAAACUCGGUUGCAAUCAAGCGCAUUGAAGGGCGGCCAUCACGUAAAUUGAGAUCAUUGUGGUGCAG___CUGCACUGGCAGCCAAGGAGUAAACGUGCCUGUUGCGGUUGAGAUUC .......(((..((((((.((.((((.......(((.((((((.((.....)).)))......((((((.....))))))))).)))..(.......))))).))))))))..))).... (-35.52 = -35.76 + 0.24)

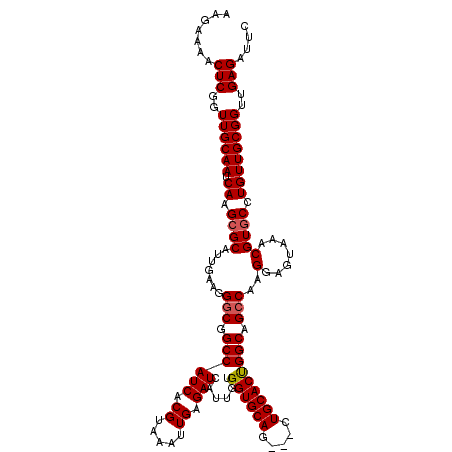
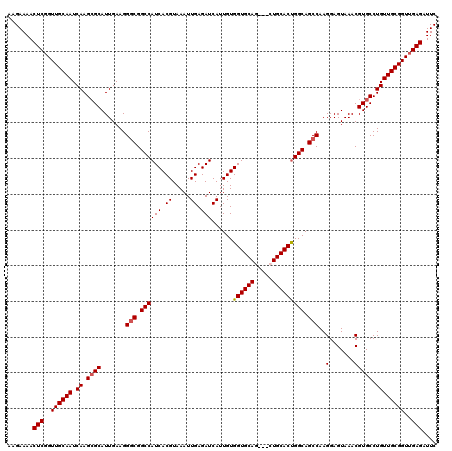
| Location | 3,234,553 – 3,234,673 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -32.82 |
| Energy contribution | -32.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3234553 120 - 22407834 GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCAGUGCAUCCACUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGCCCUUCAAUGCUCUUGAUUGCAACCGAGUUUUCUU (((.(((....((((((((..((((........(((.(((((((((.....)))))(((...(((....)))...))).)))).)))....))))..)))).))))....)))..))).. ( -28.90) >DroSec_CAF1 68649 117 - 1 GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCAGUGCAG---CUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGCCCUUCAAUGCGCUUGAUUGCAACCGAGUUUUCUU (((.(((....(((((((((.((((........(((.((((((((..---..))))(((...(((....)))...))).)))).)))....)))).))))).))))....)))..))).. ( -33.10) >DroSim_CAF1 70374 117 - 1 GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCAGUGCAG---CUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGCCCUUCAAUGCGCUUGAUUGCAACCGAGUUUUCUU (((.(((....(((((((((.((((........(((.((((((((..---..))))(((...(((....)))...))).)))).)))....)))).))))).))))....)))..))).. ( -33.10) >DroEre_CAF1 65669 117 - 1 GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCAGUGCAG---CUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGUCCUUCAAUGCGCUUGAUUGCAACAGAGUUUUCUU (((.(((....(((((((((.((((........(((.((((((((..---..))))(((...(((....)))...))).)))).)))....)))).))))).))))....)))..))).. ( -30.40) >DroYak_CAF1 64533 117 - 1 GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCGGUGCAG---UUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGCCCUUCAAUGCGCUUGAUUGCAACCGAGUUUUCUU (((.(((....(((((((((.((((........(((.((((((((..---..)))))......(((.(.......).)))))).)))....)))).))))).))))....)))..))).. ( -35.70) >consensus GAAUCUCAACCGCAACAGGCACGUUUACUCCUUGGCUGCCAGUGCAG___CUGCACCACAAUGAUCUCAAUUUACGUGAUGGCCGCCCUUCAAUGCGCUUGAUUGCAACCGAGUUUUCUU (((.(((....(((((((((.((((........(((.((((((((((...))))))(((...(((....)))...))).)))).)))....)))).))))).))))....)))..))).. (-32.82 = -32.78 + -0.04)

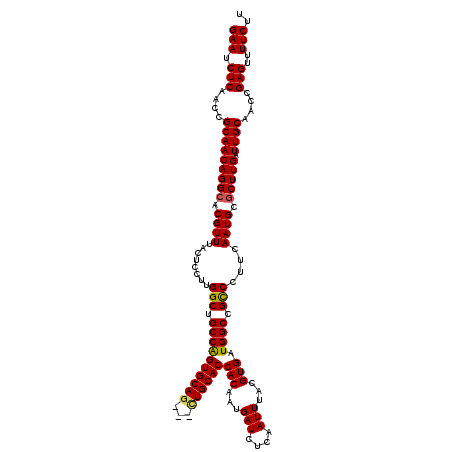
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:05 2006