
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,229,711 – 3,229,900 |
| Length | 189 |
| Max. P | 0.981410 |

| Location | 3,229,711 – 3,229,820 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -29.32 |
| Energy contribution | -29.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3229711 109 - 22407834 GUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ((.((((((((((....((((..((((...))))))))..)))))))))).))...................(((((((..........)))..))))........... ( -32.00) >DroSec_CAF1 63728 109 - 1 GUCCUGCCGCCACUACAUUGGCCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ((.((((((((((...((((((..(((...))))))))).)))))))))).))...................(((((((..........)))..))))........... ( -33.80) >DroSim_CAF1 62257 109 - 1 GUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ((.((((((((((....((((..((((...))))))))..)))))))))).))...................(((((((..........)))..))))........... ( -32.00) >DroEre_CAF1 60235 109 - 1 GUCCUGCCGCCACUCCUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUAGAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ...((((((((((....((((..((((...))))))))..))))))))))((..((....))..))......(((((((..........)))..))))........... ( -30.80) >DroYak_CAF1 59187 109 - 1 GUCCUGCCGCCACUCCUUUGGUCCUGCCCUGCAGCCAGUUGUGCUGGUGGUACAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ((..(((((((((.(..((((..((((...))))))))..).).))))))))..))................(((((((..........)))..))))........... ( -26.30) >consensus GUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCAAAGUGCCUGAACCUUGAACCAGUCGCACAAUAAAAGUAA ((.((((((((((....((((..((((...))))))))..)))))))))).))...................(((((((..........)))..))))........... (-29.32 = -29.56 + 0.24)

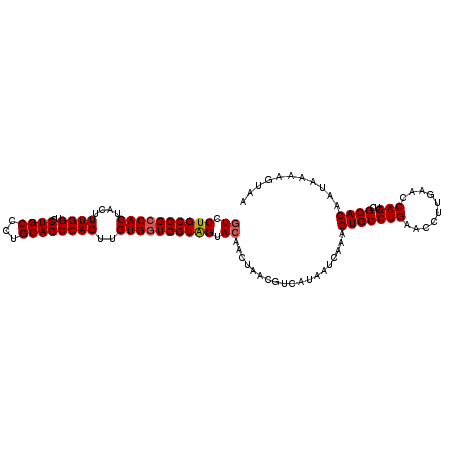
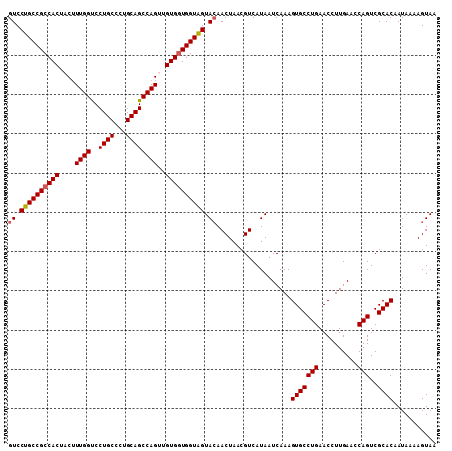
| Location | 3,229,750 – 3,229,860 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.74 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3229750 110 + 22407834 UGAUUAUGACGUUAGUUGUACUACCACCACAACUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGUUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCAC ((....((..(((((((((.........)))))))))..))...))((.(((.......((((((((((......))).))))))).((((((....))))))))))).. ( -39.10) >DroSec_CAF1 63767 110 + 1 UGAUUAUGACGUUAGUUGUACUACCACCACAACUGGCUGCAGGGCAGGGCCAAUGUAGUGGCGGCAGGACGAAGCUCCUUGCCCCUGGCACUGACACCAGUGUUGCCCAC ......((..(((((((((.........)))))))))..))(((((((((((......))))(((((((......))).)))))))(((((((....))))))))))).. ( -43.40) >DroSim_CAF1 62296 110 + 1 UGAUUAUGACGUUAGUUGUACUACCACCACAACUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCAC ((....((..(((((((((.........)))))))))..))...))((.(((.......((((((((((......))).))))))).((((((....))))))))))).. ( -39.10) >DroEre_CAF1 60274 110 + 1 UGAUUAUGACGUUAGUUCUACUACCACCACAACUGGCUGCAGGGCAGGACCAAAGGAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCAC ..............(((((.(..(((.......)))..).))))).((....(((((((..((......))..))))))).))((..((((((....))))))..))... ( -38.10) >DroYak_CAF1 59226 110 + 1 UGAUUAUGACGUUAGUUGUACCACCAGCACAACUGGCUGCAGGGCAGGACCAAAGGAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCAC ......((..(((((((((.(.....).)))))))))..)).((((.......((((((..((......))..))))))))))((..((((((....))))))..))... ( -41.91) >consensus UGAUUAUGACGUUAGUUGUACUACCACCACAACUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCAC ((....((..(((((((((.........)))))))))..))...))((.(((.......((((((((((......))).))))))).((((((....))))))))))).. (-35.14 = -35.74 + 0.60)

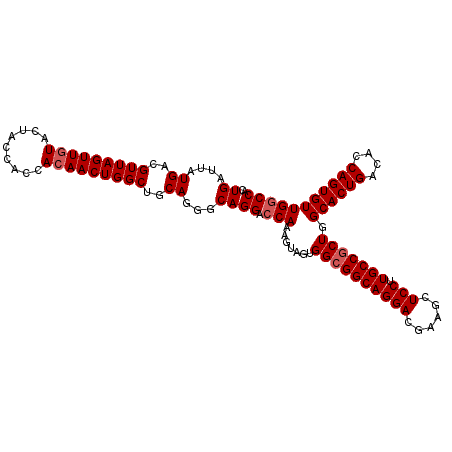
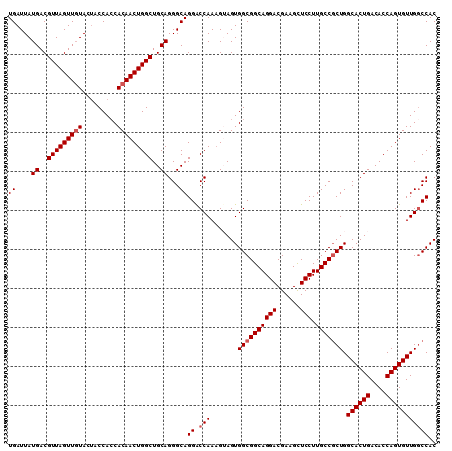
| Location | 3,229,750 – 3,229,860 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -35.56 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3229750 110 - 22407834 GUGGCCAACACUGGUGUCAGUGCCAGCGGCAAGGAACUUCGUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCA (((((.....(((((......))))).((((.(((......)))))))))))).((.(((((.((((((..(((....)))..).)))))....))))).))........ ( -39.10) >DroSec_CAF1 63767 110 - 1 GUGGGCAACACUGGUGUCAGUGCCAGGGGCAAGGAGCUUCGUCCUGCCGCCACUACAUUGGCCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCA ...(((..(((((....)))))...(((((.....)))))((.((((((((((...((((((..(((...))))))))).)))))))))).)).......)))....... ( -41.10) >DroSim_CAF1 62296 110 - 1 GUGGCCAACACUGGUGUCAGUGCCAGCGGCAAGGAGCUUCGUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCA (((((.....(((((......))))).((((.(((......)))))))))))).((.(((((.((((((..(((....)))..).)))))....))))).))........ ( -39.00) >DroEre_CAF1 60274 110 - 1 GUGGCCAACACUGGUGUCAGUGCCAGCGGCAAGGAGCUUCGUCCUGCCGCCACUCCUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUAGAACUAACGUCAUAAUCA ..(((((((((((....)))))...((((((.(((......))))))))).......))))))((((((..(((....)))..).)))))((..((....))..)).... ( -39.10) >DroYak_CAF1 59226 110 - 1 GUGGCCAACACUGGUGUCAGUGCCAGCGGCAAGGAGCUUCGUCCUGCCGCCACUCCUUUGGUCCUGCCCUGCAGCCAGUUGUGCUGGUGGUACAACUAACGUCAUAAUCA ..(((((((((((....)))))...((((((.(((......))))))))).......))))))((((...))))..((((((((.....))))))))............. ( -38.40) >consensus GUGGCCAACACUGGUGUCAGUGCCAGCGGCAAGGAGCUUCGUCCUGCCGCCACUACUUUGGUCCUGCCCUGCAGCCAGUUGUGGUGGUAGUACAACUAACGUCAUAAUCA (((((.....(((((......))))).((((.(((......)))))))))))).....(((..((((...)))))))((((((..(.(((.....))).)..)))))).. (-35.56 = -35.60 + 0.04)

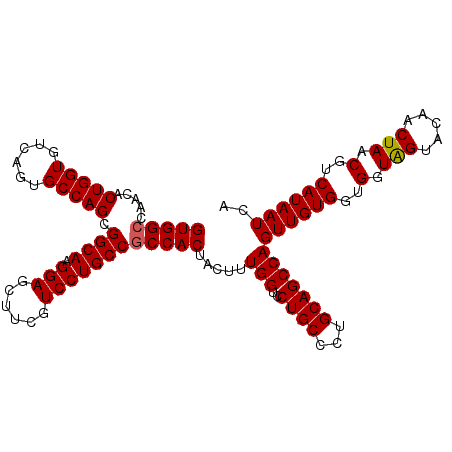

| Location | 3,229,782 – 3,229,900 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -38.16 |
| Energy contribution | -38.40 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3229782 118 + 22407834 CUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGUUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCACCAAGGCGUUGCAGAAAAAGUGGCUAAGACCUGGAUUACAA .(((((((...))))..)))..(((((((((((((((......))).)))))))..........(((((.((((((((.....(......).....)))))))).).))))))))).. ( -42.50) >DroSec_CAF1 63799 118 + 1 CUGGCUGCAGGGCAGGGCCAAUGUAGUGGCGGCAGGACGAAGCUCCUUGCCCCUGGCACUGACACCAGUGUUGCCCACCAAGGCGUUGCAGAAAAAGUGGCUAAGACCUGGAUUACAG .((((..(.(((((((((((......))))(((((((......))).)))))))(((((((....)))))))))))......((...)).......)..))))....(((.....))) ( -43.10) >DroSim_CAF1 62328 118 + 1 CUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCACCAAGGCGCUGCAGAAAAAGUGGCUAAGACAUGGAUUACAG .((((..(.((......))...((((((.((((((((......))).))))((..((((((....))))))..))......).)))))).......)..))))............... ( -46.10) >DroEre_CAF1 60306 107 + 1 CUGGCUGCAGGGCAGGACCAAAGGAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCACCAAGGGGUUGCAGAAAAAGUGGCUAAGAC----------- .((((..(...((((..((.(((((((..((......))..)))))))...((..((((((....))))))..))......))..)))).......)..))))....----------- ( -43.60) >DroYak_CAF1 59258 118 + 1 CUGGCUGCAGGGCAGGACCAAAGGAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCACCAAGGGGUUGCAGAAAAAGUGGCUAAGACCUGGAUUACAA ..(((....((......)).(((((((..((......))..))))))))))((..((((((....))))))..))..(((.(((((..(.......)..)))....)))))....... ( -44.20) >consensus CUGGCUGCAGGGCAGGACCAAAGUAGUGGCGGCAGGACGAAGCUCCUUGCCGCUGGCACUGACACCAGUGUUGGCCACCAAGGCGUUGCAGAAAAAGUGGCUAAGACCUGGAUUACAA .((((..(.((......))......(..(((((((((......))).))))((..((((((....))))))..)).........))..).......)..))))............... (-38.16 = -38.40 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:55 2006