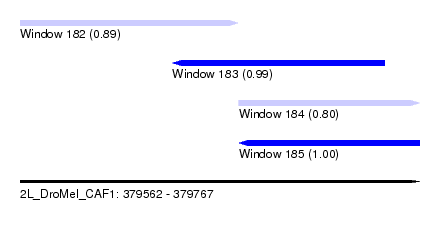
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 379,562 – 379,767 |
| Length | 205 |
| Max. P | 0.999771 |

| Location | 379,562 – 379,674 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 379562 112 + 22407834 AAUAUGUUCUCCACCGAAAUGUAGUUCAAUUCAAUUACGGCAACAUACAAUUAAAACUUGUUGCCAGAUACCUUAGAUACUG------UAUCUUGUAGAUAUACACCCGAAUUUUUGG .............((((((.((((((......))))))(((((((.............)))))))((((((..........)------)))))((((....)))).......)))))) ( -20.92) >DroSec_CAF1 47218 118 + 1 AAUAUGUUCUCCACCAAAAUGCAGUUCAAUUCAAUUACGGCAACAUACAAUUAAAACGUGUUGCCAGAUACCUUAGAUACUGUCUCGGUAUCUCGUAGAUAUACACCUGAAUUUUUGG .............((((((..(((..............((((((((...........))))))))(((((((..((((...)))).))))))).(((....)))..)))...)))))) ( -28.60) >DroSim_CAF1 47875 118 + 1 AAUAUGUUCUCCACCAAAAUGCAGUUCAAUUCAAUUACGGCAACAUACAAUUAAAACGUGUUGCCAGAUACCUUAGAUACCGUCUCGGUAUCUCGUAGAUAUACACCCGAAAUUUUGG .............(((((((..............((((((((((((...........))))))))(((((((..((((...)))).))))))).)))).............))))))) ( -26.93) >DroEre_CAF1 54180 117 + 1 AAUAUGUUCUUCGCCGA-AUGCAGUUCAAUUCAAUUACGGCAACAAACAAUUAAAACUUGUUGCCAGAUACCUCAGAUACCGUCUCGGUAUCUCGUAGAUAUACACCCGAAUUUUUGG ....(((...((...((-((........))))...(((((((((((...........))))))))(((((((..((((...)))).))))))).)))))...))).((((....)))) ( -27.00) >DroYak_CAF1 48471 117 + 1 AAUAUGUUCUCCAUCGAAAUGCAGUUCAAUUCAAUUACGGCUACAAACAAUUAAAACUUGUUGCAAGAUACCUUAGAUACUGUAUCGGUAUCUGAUAGAUAUACACCCGAAUU-UUGG ..............(((((((((((((............((.((((...........)))).))(((....))).)).))))).((((((((.....)))).....)))))))-))). ( -16.60) >consensus AAUAUGUUCUCCACCGAAAUGCAGUUCAAUUCAAUUACGGCAACAUACAAUUAAAACUUGUUGCCAGAUACCUUAGAUACUGUCUCGGUAUCUCGUAGAUAUACACCCGAAUUUUUGG .............((((((....((((...........((((((((...........))))))))..(((.((.(((((((.....)))))))...)).)))......)))))))))) (-19.56 = -20.28 + 0.72)

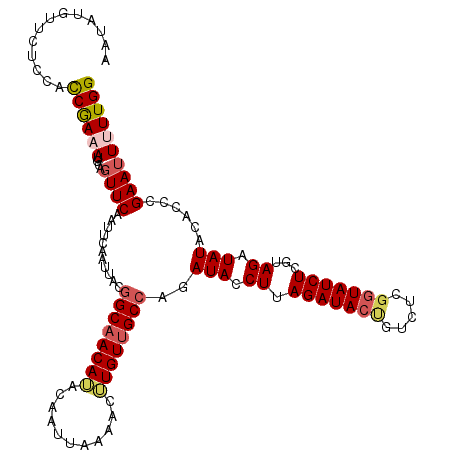
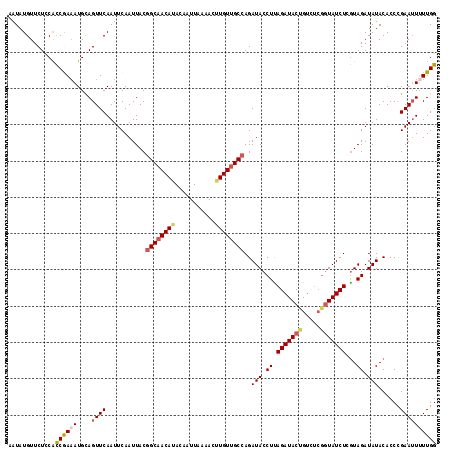
| Location | 379,640 – 379,749 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -27.97 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 379640 109 - 22407834 GCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUGCCAAAAAUUCGGGUGUAUAUCUACAAGAUA------CAGU ((((....((((.(((((((((....))((((....(((-----(((....((......)))))))))))).))))))).))))......))))...((((.....))))------.... ( -31.60) >DroSec_CAF1 47296 115 - 1 GCCCGAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUGCCAAAAAUUCAGGUGUAUAUCUACGAGAUACCGAGACAGU (((.(((.((((.(((((((((....))((((....(((-----(((....((......)))))))))))).))))))).))))...))).))).....(((.((......))))).... ( -32.00) >DroSim_CAF1 47953 115 - 1 GCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUGCCAAAAUUUCGGGUGUAUAUCUACGAGAUACCGAGACGGU (((.....((((.(((((((((....))((((....(((-----(((....((......)))))))))))).))))))).))))..((((((.((((....)))).....)))))).))) ( -35.10) >DroEre_CAF1 54257 120 - 1 GCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGUGAAUCGAAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUGCCAAAAAUUCGGGUGUAUAUCUACGAGAUACCGAGACGGU (((.....((((.((((((((((((..(((((.....(((......)))..((......)))))))))))).))))))).))))...(((((.((((....)))).....)))))..))) ( -35.60) >DroYak_CAF1 48549 119 - 1 GCCCCAAAUUGGGCGCUGUCGAGCAAUCGAGCAACAACGAUUGAAAUCAAAUCAAAUCAGAGUUCAUGUUCAGAUAGUUGCCAA-AAUUCGGGUGUAUAUCUAUCAGAUACCGAUACAGU ((((......))))((((((((....))))(((((...((((.......))))..(((.(((......))).))).)))))...-...((((.....((((.....))))))))..)))) ( -28.90) >consensus GCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG_____AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUGCCAAAAAUUCGGGUGUAUAUCUACGAGAUACCGAGACAGU ((((....((((.((((((((((((..(((((.............................)))))))))).))))))).))))......))))...((((.....)))).......... (-27.97 = -28.25 + 0.28)

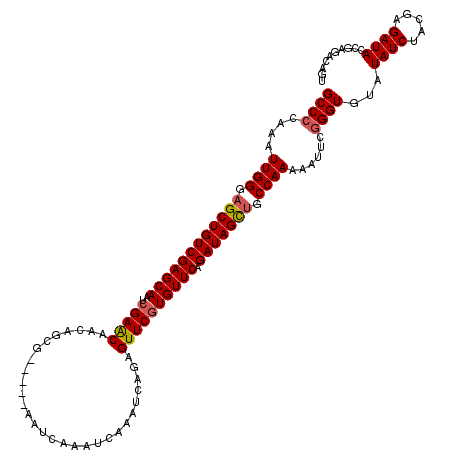
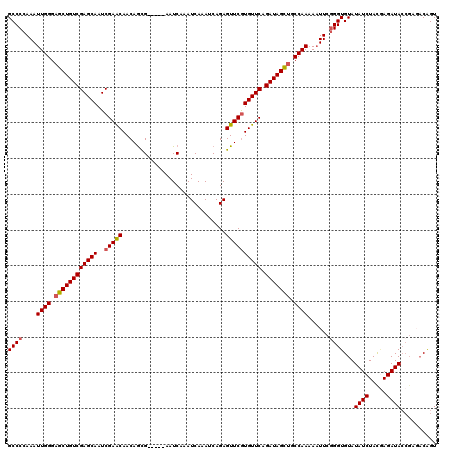
| Location | 379,674 – 379,767 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 379674 93 + 22407834 CAGCUAUCUGAACACGAACUCUGAUUUGAUUUGAUU-----CGCUGUUGUUCGAUUGCUCGACAGCUCCCAAUUUGGGGCGCUGUCUUUUGAAGGGAU .(((.(((.(((((((((.((...........))))-----))....)))))))).))).((((((((((.....)))).))))))............ ( -27.00) >DroSec_CAF1 47336 93 + 1 CAGCUAUCUGAACACGAACUCUGAUUUGAUUUGAUU-----CGCUGUUGUUCGAUUGCUCGACAGCUCCCAAUUUCGGGCGCUGUCUUUUGAAGGGAU .(((.(((.(((((((((.((...........))))-----))....)))))))).))).((((((.(((......))).))))))............ ( -27.10) >DroSim_CAF1 47993 93 + 1 CAGCUAUCUGAACACGAACUCUGAUUUGAUUUGAUU-----CGCUGUUGUUCGAUUGCUCGACAGCUCCCAAUUUGGGGCGCUGUCUUUUGAAGGGAU .(((.(((.(((((((((.((...........))))-----))....)))))))).))).((((((((((.....)))).))))))............ ( -27.00) >DroEre_CAF1 54297 98 + 1 CAGCUAUCUGAACACGAACUCUGAUUUGAUUUGAUUCGAUUCACUGUUGUUCGAUUGCUCGACAGCUCCCAAUUUGGGGCGCUGUCUUUUGAUGGGCU ...(((((((((((...((..(((.((((......)))).)))..)))))))).......((((((((((.....)))).))))))....)))))... ( -31.60) >DroYak_CAF1 48588 98 + 1 CAACUAUCUGAACAUGAACUCUGAUUUGAUUUGAUUUCAAUCGUUGUUGCUCGAUUGCUCGACAGCGCCCAAUUUGGGGCGCUGUCUUUUGAAGGGCU ((((.((.((((.((.((.((......)).)).)))))))).))))..((((((....))((((((((((......)))))))))).......)))). ( -31.80) >consensus CAGCUAUCUGAACACGAACUCUGAUUUGAUUUGAUU_____CGCUGUUGUUCGAUUGCUCGACAGCUCCCAAUUUGGGGCGCUGUCUUUUGAAGGGAU .(((.(((.(((((((......((((......))))........)).)))))))).))).((((((.(((......))).))))))............ (-21.98 = -22.22 + 0.24)

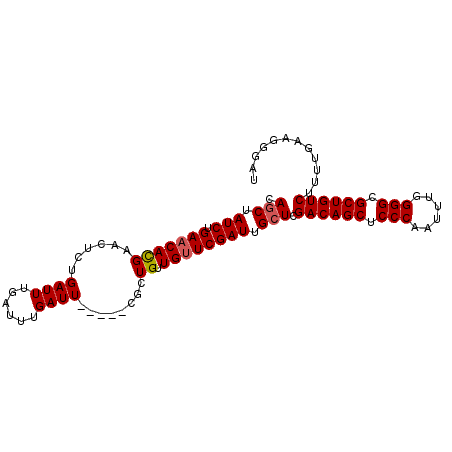
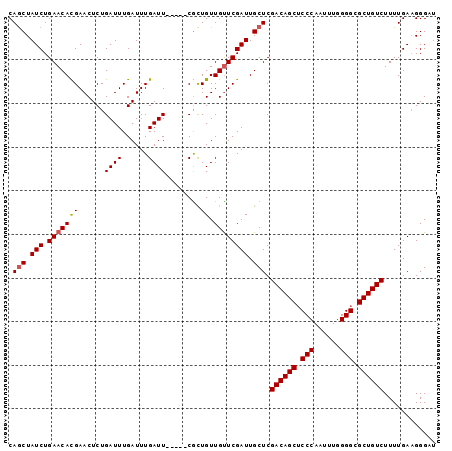
| Location | 379,674 – 379,767 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -25.20 |
| Energy contribution | -24.56 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 379674 93 - 22407834 AUCCCUUCAAAAGACAGCGCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUG ............((((((..(((.....))).)))))).(((.(((((((....(((-----(((....((......)))))))))))).))).))). ( -28.30) >DroSec_CAF1 47336 93 - 1 AUCCCUUCAAAAGACAGCGCCCGAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUG ............((((((.((((....)))).)))))).(((.(((((((....(((-----(((....((......)))))))))))).))).))). ( -28.90) >DroSim_CAF1 47993 93 - 1 AUCCCUUCAAAAGACAGCGCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG-----AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUG ............((((((..(((.....))).)))))).(((.(((((((....(((-----(((....((......)))))))))))).))).))). ( -28.30) >DroEre_CAF1 54297 98 - 1 AGCCCAUCAAAAGACAGCGCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGUGAAUCGAAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUG (((..(((....((((((..(((.....))).))))))(((((..(((((.....(((......)))..((......)))))))))))).))).))). ( -30.80) >DroYak_CAF1 48588 98 - 1 AGCCCUUCAAAAGACAGCGCCCCAAAUUGGGCGCUGUCGAGCAAUCGAGCAACAACGAUUGAAAUCAAAUCAAAUCAGAGUUCAUGUUCAGAUAGUUG .((.(.......((((((((((......))))))))))((....))).))..((((..(((((......((......)).......)))))...)))) ( -31.02) >consensus AUCCCUUCAAAAGACAGCGCCCCAAAUUGGGAGCUGUCGAGCAAUCGAACAACAGCG_____AAUCAAAUCAAAUCAGAGUUCGUGUUCAGAUAGCUG ............((((((.(((......))).)))))).(((.((((((((..(((.......................)))..))))).))).))). (-25.20 = -24.56 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:30 2006