
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,224,132 – 3,224,225 |
| Length | 93 |
| Max. P | 0.987473 |

| Location | 3,224,132 – 3,224,225 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.88 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.25 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
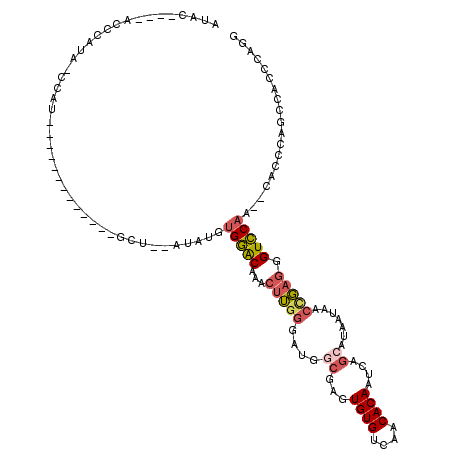
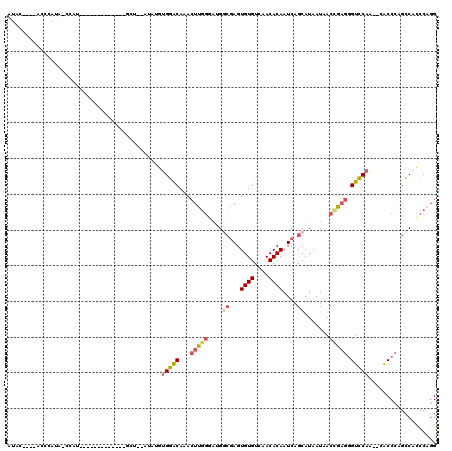
>2L_DroMel_CAF1 3224132 93 + 22407834 CCUGGGUGGCUGGGUG--UUGGACCCUCGGUUAUUAUGCUGAUUGUGUUGACACACUCGCCAUCCCAAGUAUGUCCACAUAU--ACC----------------GAUAUAG-------UAU ..((((((((.(((((--(..(((..(((((......)))))..)).)..)))).)).)))).)))).(((((....)))))--...----------------.......-------... ( -30.60) >DroSec_CAF1 58173 99 + 1 CCUGGGUGGCUGGGUG--UUGGACCCUCGGUUAUUAUGCUGAUUGUGUUGACACACUCGCCAUCCCAAGUAUGCCCACAUAU--AGC-------------AUGGCUAUGGGU----AUAU ..((((((((.(((((--(..(((..(((((......)))))..)).)..)))).)).)))).)))).(((((((((....(--(((-------------...)))))))))----)))) ( -40.20) >DroSim_CAF1 56677 92 + 1 CCUGGGUGGCUGGGUG--UUGGACCCUCGGUUAUUAUGCUGAUUGUGUUGACACACUCGCCAUUCCAAGUAUGUCCAC----------------------AUGGCUAUGGGU----AUAU ..((((((((.(((((--(..(((..(((((......)))))..)).)..)))).)).)))).)))).((((..(((.----------------------.......)))..----)))) ( -31.00) >DroEre_CAF1 54765 117 + 1 CCAGGGUGGCUGGGUG--UUGGACCCUCAGUUAUUAUGCUGAUUGUGUUGACACACUCGCCCUCCCAAGUUUGUCCAGAUAUACAGCCAGCAUAUAUAGCAUAG-UAUAGGUACAUGUAU ....(.((((((..((--((((((..(((((......))))).((((....)))).................))))).)))..)))))).).......((((.(-((....))))))).. ( -32.50) >DroYak_CAF1 53515 104 + 1 CCUGGGCGGCUGGGUG--UUGGACCCUCAGUUAUUAUGCUGAUUGUGUUGACACACUCGGCCACCCAACUUUGCCCACAAAU--AGCC-------AUAGUAUAG-UAU----ACUUGUAU ..((((.(((((((((--((.((((.(((((......)))))..).))).).)))))))))).))))(((.(((........--....-------...))).))-)..----........ ( -32.66) >DroPer_CAF1 68205 94 + 1 CUUGGAGGUCACGGCCAUAUGUACUCUAGGUUAUUAUGCUGAUUGUGUUGACACACUGUGCGUCUGUUGUUUGUACUU----------------------UCGGUGCUGACC----UUAG ....(((((((((((...((((((.....((((..((((.....)))))))).....))))))..)))))..((((..----------------------...)))).))))----)).. ( -24.10) >consensus CCUGGGUGGCUGGGUG__UUGGACCCUCGGUUAUUAUGCUGAUUGUGUUGACACACUCGCCAUCCCAAGUAUGUCCACAUAU__AGC_____________AUGG_UAUGGGU____AUAU ...(((.(((.(((((....(.((..(((((......)))))..)).).....)))))))).)))....................................................... (-15.36 = -15.25 + -0.11)



| Location | 3,224,132 – 3,224,225 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.88 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3224132 93 - 22407834 AUA-------CUAUAUC----------------GGU--AUAUGUGGACAUACUUGGGAUGGCGAGUGUGUCAACACAAUCAGCAUAAUAACCGAGGGUCCAA--CACCCAGCCACCCAGG ...-------((((((.----------------...--..)))))).....((((((.((((((.((((....)))).))..............((((....--.)))).)))))))))) ( -29.80) >DroSec_CAF1 58173 99 - 1 AUAU----ACCCAUAGCCAU-------------GCU--AUAUGUGGGCAUACUUGGGAUGGCGAGUGUGUCAACACAAUCAGCAUAAUAACCGAGGGUCCAA--CACCCAGCCACCCAGG .(((----.((((((.....-------------...--...)))))).)))((((((.((((((.((((....)))).))..............((((....--.)))).)))))))))) ( -34.20) >DroSim_CAF1 56677 92 - 1 AUAU----ACCCAUAGCCAU----------------------GUGGACAUACUUGGAAUGGCGAGUGUGUCAACACAAUCAGCAUAAUAACCGAGGGUCCAA--CACCCAGCCACCCAGG ....----..(((((....)----------------------)))).....(((((..((((((.((((....)))).))..............((((....--.)))).)))).))))) ( -26.90) >DroEre_CAF1 54765 117 - 1 AUACAUGUACCUAUA-CUAUGCUAUAUAUGCUGGCUGUAUAUCUGGACAAACUUGGGAGGGCGAGUGUGUCAACACAAUCAGCAUAAUAACUGAGGGUCCAA--CACCCAGCCACCCUGG .....(((.((((((-(...((((.......)))).)))))...)))))...(..((..(((..((((....))))..((((........))))((((....--.)))).)))..))..) ( -31.70) >DroYak_CAF1 53515 104 - 1 AUACAAGU----AUA-CUAUACUAU-------GGCU--AUUUGUGGGCAAAGUUGGGUGGCCGAGUGUGUCAACACAAUCAGCAUAAUAACUGAGGGUCCAA--CACCCAGCCGCCCAGG ..((((((----(..-(........-------)..)--))))))((((...(((((((((((..((((....))))..((((........)))).)))....--)))))))).))))... ( -35.30) >DroPer_CAF1 68205 94 - 1 CUAA----GGUCAGCACCGA----------------------AAGUACAAACAACAGACGCACAGUGUGUCAACACAAUCAGCAUAAUAACCUAGAGUACAUAUGGCCGUGACCUCCAAG ...(----(((((...(((.----------------------..((((........(((((.....))))).......((((.........)).))))))...)))...))))))..... ( -18.60) >consensus AUAC____ACCCAUA_CCAU_____________GCU__AUAUGUGGACAAACUUGGGAUGGCGAGUGUGUCAACACAAUCAGCAUAAUAACCGAGGGUCCAA__CACCCAGCCACCCAGG ...........................................(((((...(((((....((...((((....))))....)).......))))).)))))................... (-13.96 = -14.35 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:43 2006