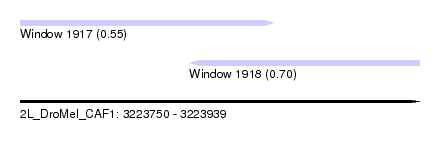
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,223,750 – 3,223,939 |
| Length | 189 |
| Max. P | 0.699518 |

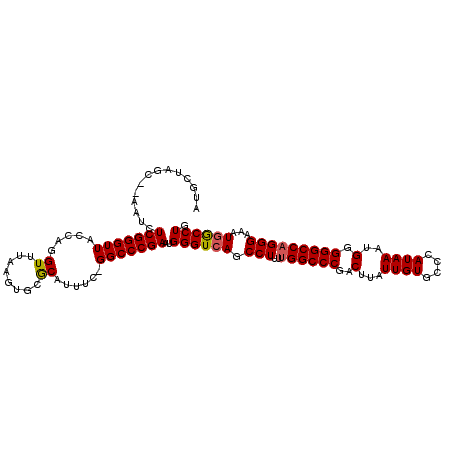
| Location | 3,223,750 – 3,223,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3223750 120 + 22407834 UUUCGGUCCUUUGGCAAGAUUAAGAUUUAAAUAAUUUAUUUGAAGUACAUUUAAUUUGCUGGCAACACACAAAGAUAUCCCAGGCCAUUUCCCUGGCCCUCAUUUAUGGGCACAAUAAGU ....((..((((((((.(((((((.((((((((...)))))))).....))))))))))((....))..)))))....))((((.......))))((((........))))......... ( -25.30) >DroSec_CAF1 57794 120 + 1 UUUCGGUCCUUUGGCAAGAUUAAGAUUUAAAUAAUUUAUUUGAAGUACAUUUAAUUUGCUGGCAACACACAAAGAUAUCCCAGGCCAUUUCCCUGGCCCCCAUUUAUGGGCACAAUAAGU ....((..((((((((.(((((((.((((((((...)))))))).....))))))))))((....))..)))))....))..(((((......)))))((((....)))).......... ( -25.40) >DroSim_CAF1 56297 119 + 1 UUUCGGUCCUUUGCCAAGAUUAAGAUUUAAAUAAUUUAUUUGAAGUACAUUUAAUUUGCUGGCAACACACAAAGAUAUCCCAGGCCAU-UCCCUGGCCCCCAUUUAUGGGCACAAUAAGU .....((((.((((((((((((((.((((((((...)))))))).....))))))))..)))))).................(((((.-....))))).........))))......... ( -25.20) >DroEre_CAF1 54393 117 + 1 UU--GGCCCGUUGGCAAGAUUAAGAUUUAAAUAAUUUAGUUGAAGUACAUUUAAUUUGCUGGCAACACACAAAGAUAUUCCAGGUCAUUUCCC-GGCCCCCAUUUAUGGGCACAAUAAGU ((--(((((((..((((.....(((((.....))))).((((((.....))))))))))..))...................((((.......-)))).........)))).)))..... ( -24.40) >DroYak_CAF1 53133 118 + 1 UU--GGCCCGUUGGCAAGAUUAAGAUUUAAAUAAUUUAUUUGAAGUACAUUUAAUUUGCUGGCAACACACAAUGAUAUCCCAGGCUAUUUCCCUGGCCCUCAUUUAUGGGCACAAUAAGU ((--(((((((..(((.(((((((.((((((((...)))))))).....))))))))))..)).......(((((....(((((.......)))))...)))))...)))).)))..... ( -31.60) >consensus UUUCGGUCCUUUGGCAAGAUUAAGAUUUAAAUAAUUUAUUUGAAGUACAUUUAAUUUGCUGGCAACACACAAAGAUAUCCCAGGCCAUUUCCCUGGCCCCCAUUUAUGGGCACAAUAAGU .....(((((((((((.(((((((.((((((((...)))))))).....))))))))))((....))..)))).........(((((......))))).........))))......... (-21.06 = -21.06 + 0.00)

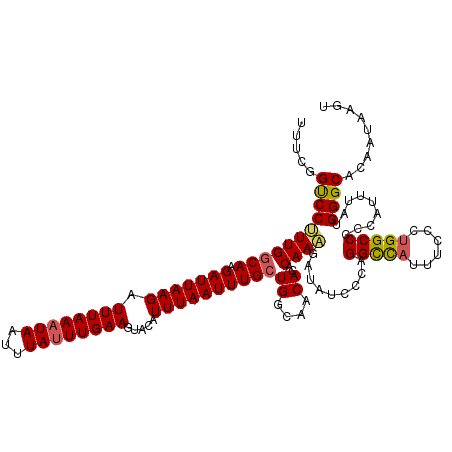
| Location | 3,223,830 – 3,223,939 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -39.76 |
| Consensus MFE | -31.62 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3223830 109 - 22407834 AUGCUAGC--AAUCUCGGGUUACCAGGUUUAAGUGCACAUUUC-GGCCCGAAUGGGUCAACCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGAGGGCCAGGGAAAUGGCCUG (((...((--(((.((((((((..(((((.((((....)))).-(((((....))))))))))..))))))))...)))))...)))......((((((......)))))). ( -37.80) >DroSec_CAF1 57874 109 - 1 AUGCUAGC--AAUCUCGGGUUACCAGGUUUAAGUGCGCAUUUC-GGCCCGAAUGGGUCAGCCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGGGGGCCAGGGAAAUGGCCUG ......((--(((.((((((((..(((((...(....).....-(((((....))))))))))..))))))))...))))).((((....))))(((((......))))).. ( -40.20) >DroSim_CAF1 56377 110 - 1 AUGCUAGCACAAUCUCGGGUUACCAGGUUUAAGUGCGCAUUUC-GGCCCGAAUGGGUCAGCCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGGGGGCCAGGGA-AUGGCCUG ......(((((((.((((((((..(((((...(....).....-(((((....))))))))))..))))))))...)))))))(((....)))((((((....-.)))))). ( -44.00) >DroEre_CAF1 54471 107 - 1 AUCCUAGC--AAUCUCGGGUUACC-GGUUUAAGUGCGCAUUUC-GGCCCGAAUGGGUCAGCCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGGGGGCC-GGGAAAUGACCUG .(((..((--(...((((....))-))......)))......(-(((((.((((((((.(((....)))..))))))))....(((....))))))))-))))......... ( -38.20) >DroYak_CAF1 53211 109 - 1 AUCCUAGC--AAUCUCGGGUUACC-GGUUUAAGUGCGCAUUUUGGGCCCGAAUGGGUCAGCCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGAGGGCCAGGGAAAUAGCCUG .((((.((--(...((((....))-))......))).((((((((((.(.((((((((.(((....)))..))))))))).))))).))))))))).((((.......)))) ( -38.60) >consensus AUGCUAGC__AAUCUCGGGUUACCAGGUUUAAGUGCGCAUUUC_GGCCCGAAUGGGUCAGCCUUUUGGCCCGACUUAUUGUGCCCAUAAAUGGGGGCCAGGGAAAUGGCCUG ..............(((((((.....((........))......)))))))..((((((.(((..((((((..(...((((....))))..).)))))))))...)))))). (-31.62 = -31.70 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:41 2006