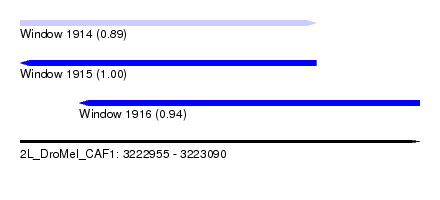
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,222,955 – 3,223,090 |
| Length | 135 |
| Max. P | 0.999755 |

| Location | 3,222,955 – 3,223,055 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3222955 100 + 22407834 GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGUGAGCAAUUCGGUGGCUAAACUGAUUUUAGCCAUUUGGCGUGCUCUGGGCCAAAAUAGAUGUAGACGAAG .(((((((........)))....((((((.((((((..((((((((((((.....)))))))).))))..))))).).))))))......))))...... ( -28.10) >DroSec_CAF1 57039 99 + 1 GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGUGAGCAGUUCGGUGGCUAAACUGAUUUUAGCCAUUUGGCGUGCUCUGGGCCAAAAUAGAUGUAGAC-AAG .(((((((........)))....((((((.(((((((((..(((((((((.....)))))))))..))).))))).).))))))......))))..-... ( -31.30) >DroSim_CAF1 55500 99 + 1 GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGUGAGCAAUUCGGUGGCUAAACUGAUUUUAGCCAUUUGGCGUGCUCUGGGCCAAAAUAGAUGUAGAC-AAG .(((((((........)))....((((((.((((((..((((((((((((.....)))))))).))))..))))).).))))))......))))..-... ( -28.10) >DroEre_CAF1 53557 95 + 1 GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGAGAGCAACUGGGUGGCUAAACUGAUUUUAGCCAUUUGGCGAGCUCUGGGCCAAAAUA----UAGAC-GCG ((.(((.....))).........((((((.(((((..(..((((((((((.....))))))))))..)...))))).).)))))...----.....-)). ( -29.60) >DroYak_CAF1 52249 93 + 1 GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGUGAGCAAUUCGGUGGCUAAACUGAUUUUAGCCAUUUGGCGUGCUCUGGGCCAAAAU------GAAC-GUG ((.(((.....))).))......((((((.((((((..((((((((((((.....)))))))).))))..))))).).))))))..------....-... ( -26.50) >consensus GCUGCUCGAGAAGUUGUGAAUUAUUUGGUGUGAGCAAUUCGGUGGCUAAACUGAUUUUAGCCAUUUGGCGUGCUCUGGGCCAAAAUAGAUGUAGAC_AAG ((.(((.....))).))......((((((..(((((....((((((((((.....)))))))))).....)))))...))))))................ (-24.86 = -25.06 + 0.20)


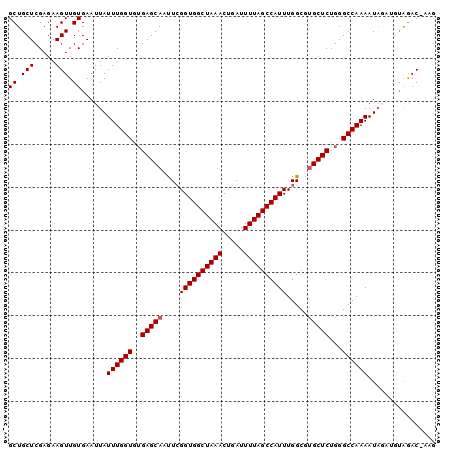
| Location | 3,222,955 – 3,223,055 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3222955 100 - 22407834 CUUCGUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAAUUCACAACUUCUCGAGCAGC .((((...........((((((.........))))))((((((((.....)))))))).))))((((((........................)))))). ( -21.56) >DroSec_CAF1 57039 99 - 1 CUU-GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAACUGCUCACACCAAAUAAUUCACAACUUCUCGAGCAGC ...-............((((((.........))))))((((((((.....)))))))).....((((((........................)))))). ( -21.46) >DroSim_CAF1 55500 99 - 1 CUU-GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAAUUCACAACUUCUCGAGCAGC ...-............((((((.........))))))((((((((.....)))))))).....((((((........................)))))). ( -19.36) >DroEre_CAF1 53557 95 - 1 CGC-GUCUA----UAUUUUGGCCCAGAGCUCGCCAAAUGGCUAAAAUCAGUUUAGCCACCCAGUUGCUCUCACCAAAUAAUUCACAACUUCUCGAGCAGC ...-.....----...((((((.........))))))((((((((.....))))))))....(((((((........................))))))) ( -21.66) >DroYak_CAF1 52249 93 - 1 CAC-GUUC------AUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAAUUCACAACUUCUCGAGCAGC ...-((((------..((((((.........))))))((((((((.....))))))))..))))(((((........................))))).. ( -20.96) >consensus CUU_GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAAUUCACAACUUCUCGAGCAGC ................((((((.........))))))((((((((.....)))))))).....((((((........................)))))). (-19.90 = -19.74 + -0.16)

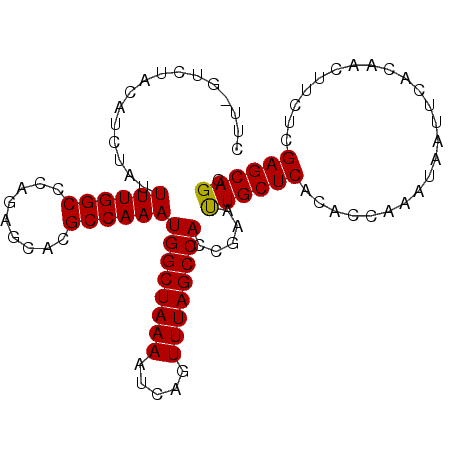
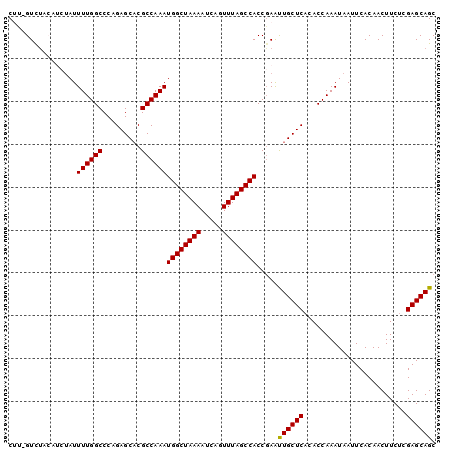
| Location | 3,222,975 – 3,223,090 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.52 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3222975 115 - 22407834 CAUAAUCCGUC--GAGUGCACUGAACUCCGGC-AUCUCCUUCGUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAA ........(((--((((.......))).))))-.....................(((((....(((((.......((((((((.....))))))))......)))))...)))))... ( -25.62) >DroSec_CAF1 57059 114 - 1 CAUAAUCCGUC--GGGUGCACUGAACUCCGGC-AUCUCCUU-GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAACUGCUCACACCAAAUAA ........(((--((((.......)).)))))-........-............(((((....(((((.......((((((((.....))))))))......)))))...)))))... ( -27.72) >DroSim_CAF1 55520 114 - 1 CAUAAUCCGUC--GGGUGCACUGAACUCCGGC-ACCUCCUU-GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAA ........(((--((((.......)).)))))-........-............(((((....(((((.......((((((((.....))))))))......)))))...)))))... ( -27.72) >DroEre_CAF1 53577 113 - 1 CAUAAUCCUGCAAGAGUGCACUGAACUCCGGCCAUCUUCGC-GUCUA----UAUUUUGGCCCAGAGCUCGCCAAAUGGCUAAAAUCAGUUUAGCCACCCAGUUGCUCUCACCAAAUAA .............(((.((((((..(((.(((((.......-.....----.....)))))..))).........((((((((.....))))))))..))).))).)))......... ( -29.93) >DroYak_CAF1 52269 109 - 1 CAUAAUCCUGC--GAGUGCACUGAACUCCGGCCAUCACCAC-GUUC------AUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAA .........((--((((((.(((......(((((.......-....------....)))))))).))))......((((((((.....)))))))).....))))............. ( -27.56) >consensus CAUAAUCCGUC__GAGUGCACUGAACUCCGGC_AUCUCCUU_GUCUACAUCUAUUUUGGCCCAGAGCACGCCAAAUGGCUAAAAUCAGUUUAGCCACCGAAUUGCUCACACCAAAUAA ......(((....((((.......))))))).......................(((((....(((((.......((((((((.....))))))))......)))))...)))))... (-21.28 = -21.64 + 0.36)

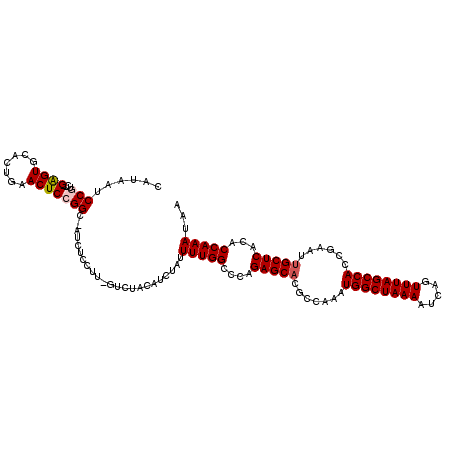
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:39 2006