| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,221,438 – 3,221,538 |
| Length | 100 |
| Max. P | 0.558798 |

| Location | 3,221,438 – 3,221,538 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
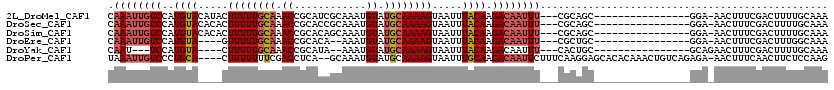
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
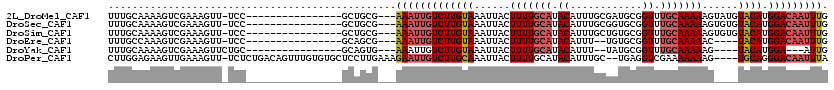
>2L_DroMel_CAF1 3221438 100 + 22407834 UUUGCAAAAGUCGAAAGUU-UCC----------------GCUGCG---AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUUGCGAUGCGGUUUGCAAAAAGUAUGUACAUGGACAAUUUG ..((((.....(....)..-...----------------..))))---(((((((((((((...((((((((((......)))..(((.....))))))))))..)))).))))))))). ( -25.70) >DroSec_CAF1 55529 100 + 1 UUUGCAAAAGUCGAAAGUU-UCC----------------GCUGCG---AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUUGCGGUGCGGUUUGCAAAAAGUGUGUACAUGGACAAUUUG ..((((.....(....)..-...----------------..))))---(((((((((((((...((((((((((......)))..(((.....))))))))))..)))).))))))))). ( -25.30) >DroSim_CAF1 54012 100 + 1 UUUGCAAAAGUCGAAAGUU-UCC----------------GCUGCG---AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUUGCUGUGCGGUUUGCAAAAAGUGUGUACAUGGACAAUUUG ..((((.....(....)..-...----------------..))))---(((((((((((((...((((((((((.((....((...)).)).))).)))))))..)))).))))))))). ( -25.00) >DroEre_CAF1 52054 94 + 1 UUUGCCAAAGUCGAAAGUU-UCC----------------GCAGCG---AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUU--UGUGCGGUUUGCAAAAAC----UACAUGGACAAUUUG .((((((.(((.....((.-.((----------------(((.((---((((.((..((((((.....)))))).))))))--)))))))...))....))----)...))).))).... ( -21.60) >DroYak_CAF1 50809 92 + 1 UUUGCAAAAGUCGAAAGUUCUGC----------------GCAGUG---AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUU--UAUGCGGUUUGCAAAAAG----UACAUGGA---AUUG ((((((((((.(....)..)).(----------------(((.((---((((.((..((((((.....)))))).))))))--)))))).))))))))...----........---.... ( -20.90) >DroPer_CAF1 65961 113 + 1 CUUGGAGAAGUUGAAAGUU-UCUCUGACAGUUUGUGUGCUCCUUGAAAGAAUUGUCUUGCAAAUUACUUUUGCAUACAUUUGC--UGAGGUCGAAAAAAAG----UGCAGGGACAAUUUA .(..((((((.......))-))))..)..((((.((..(((((((...((((.((..((((((.....)))))).))))))..--))))).........))----..)).))))...... ( -26.30) >consensus UUUGCAAAAGUCGAAAGUU_UCC________________GCUGCG___AAAUUGUCUUGUAAAUUACUUUUGCAUACAUUUGCUGUGCGGUUUGCAAAAAG____UACAUGGACAAUUUG ................................................(((((((((((((......(((((((.((............)).)))))))......)))).))))))))). (-14.90 = -15.65 + 0.75)
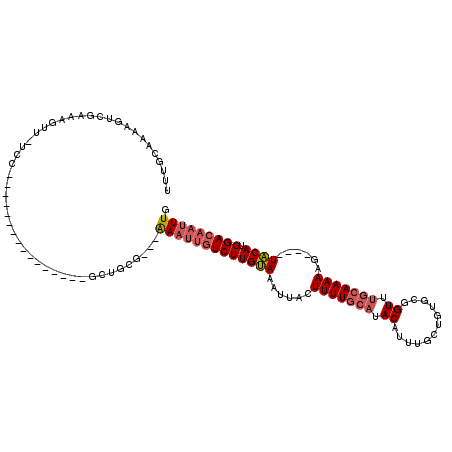
| Location | 3,221,438 – 3,221,538 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3221438 100 - 22407834 CAAAUUGUCCAUGUACAUACUUUUUGCAAACCGCAUCGCAAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU---CGCAGC----------------GGA-AACUUUCGACUUUUGCAAA .((((((((..((((..(((((((.(((....((((.....)))).))))))))))...)))).))))))))---.(((((----------------(((-....))))....))))... ( -21.00) >DroSec_CAF1 55529 100 - 1 CAAAUUGUCCAUGUACACACUUUUUGCAAACCGCACCGCAAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU---CGCAGC----------------GGA-AACUUUCGACUUUUGCAAA .((((((((..((((...(((((((((.....)))..(((......)))))))))....)))).))))))))---.(((((----------------(((-....))))....))))... ( -19.20) >DroSim_CAF1 54012 100 - 1 CAAAUUGUCCAUGUACACACUUUUUGCAAACCGCACAGCAAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU---CGCAGC----------------GGA-AACUUUCGACUUUUGCAAA .((((((((..((((...(((((((((.....)))..(((......)))))))))....)))).))))))))---.(((((----------------(((-....))))....))))... ( -19.10) >DroEre_CAF1 52054 94 - 1 CAAAUUGUCCAUGUA----GUUUUUGCAAACCGCACA--AAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU---CGCUGC----------------GGA-AACUUUCGACUUUGGCAAA .((((((((..((((----(((((((((.((......--....)).))))))))....))))).))))))))---.(((.(----------------(((-....)))).....)))... ( -18.60) >DroYak_CAF1 50809 92 - 1 CAAU---UCCAUGUA----CUUUUUGCAAACCGCAUA--AAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU---CACUGC----------------GCAGAACUUUCGACUUUUGCAAA ....---.....(((----((((((((.....))).)--))).))))((((((((.........((.....)---).((..----------------..))........))))))))... ( -13.20) >DroPer_CAF1 65961 113 - 1 UAAAUUGUCCCUGCA----CUUUUUUUCGACCUCA--GCAAAUGUAUGCAAAAGUAAUUUGCAAGACAAUUCUUUCAAGGAGCACACAAACUGUCAGAGA-AACUUUCAACUUCUCCAAG ..(((((((..(((.----................--(((......))).(((....)))))).))))))).......((((.(((.....)))..((((-...)))).....))))... ( -15.60) >consensus CAAAUUGUCCAUGUA____CUUUUUGCAAACCGCACAGCAAAUGUAUGCAAAAGUAAUUUACAAGACAAUUU___CGCAGC________________GGA_AACUUUCGACUUUUGCAAA .((((((((..((((.....((((((((.((............)).)))))))).....)))).))))))))................................................ (-11.93 = -12.68 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:37 2006