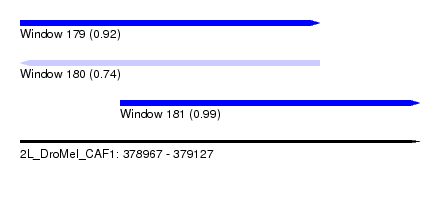
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 378,967 – 379,127 |
| Length | 160 |
| Max. P | 0.985437 |

| Location | 378,967 – 379,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 378967 120 + 22407834 AAUUUUAUUAACUCAAUUUUGAGGGUCGAAAGAUUUCGCCUUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCA ....................(((((.((((....)))))))))......(((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))). ( -25.40) >DroSec_CAF1 46605 120 + 1 AAAUUUAUUAACUCAAUUUUGAGGGGCGAAAGAUUUCGCCUUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCG ...........((((....))))(((((((....)))))))........(((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))). ( -30.00) >DroSim_CAF1 47267 120 + 1 AAUUUUAUUAACUCAAUUUUGAGGGGCGAAAGAUUUCGCCUUUGCCUUAGCUGAUAGUACGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCG ...........((((....))))(((((((....)))))))........(((((((((.(((((....)))))((...((((.((.....)).))))....)).......))))))))). ( -30.10) >DroEre_CAF1 53581 120 + 1 AACUUUAUUAACUCAAUUUUGAGGAGCACAAGAUUUCGCCUUUGCCUUUGCUGAUAGUUCGAUAAGUAUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCG ...........((((....))))..(((.(((.......))))))....(((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))). ( -22.80) >DroYak_CAF1 47819 120 + 1 AACUUUAUUAACUCAACUCCGAGGGGCGAAAGAUUUCGCCUUCGCCUGAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCG ...........(((......)))(((((((.(......).)))))))..(((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))). ( -30.00) >consensus AACUUUAUUAACUCAAUUUUGAGGGGCGAAAGAUUUCGCCUUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCG ...........(((......)))(((((((....)))))))........(((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))). (-24.76 = -25.76 + 1.00)

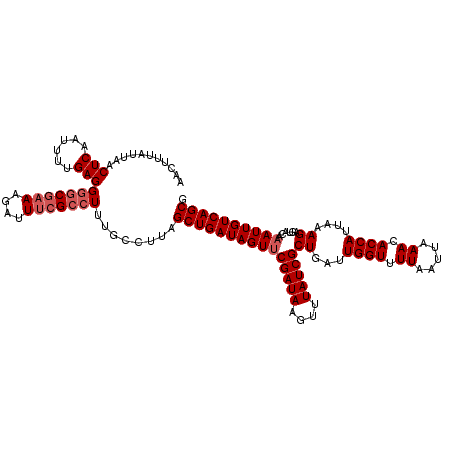
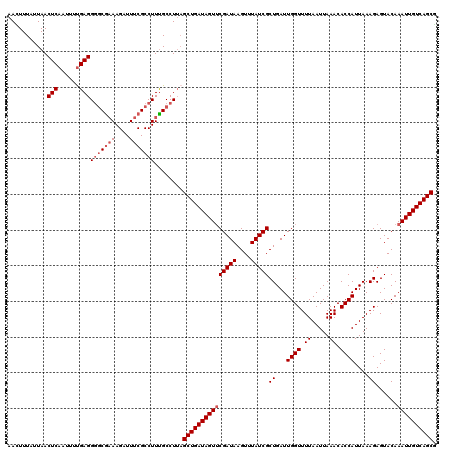
| Location | 378,967 – 379,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -21.02 |
| Energy contribution | -20.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 378967 120 - 22407834 UGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAAACUUAUCGAACUAUCAGCUAAGGCAAAGGCGAAAUCUUUCGACCCUCAAAAUUGAGUUAAUAAAAUU .(((((.................((((...........)))).....(((((....))))).....))))).........((((((....)))).))((((....))))........... ( -19.60) >DroSec_CAF1 46605 120 - 1 CGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAAACUUAUCGAACUAUCAGCUAAGGCAAAGGCGAAAUCUUUCGCCCCUCAAAAUUGAGUUAAUAAAUUU .(((((.................((((...........)))).....(((((....))))).....))))).........((((((....)))))).((((....))))........... ( -23.70) >DroSim_CAF1 47267 120 - 1 CGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAAACUUAUCGUACUAUCAGCUAAGGCAAAGGCGAAAUCUUUCGCCCCUCAAAAUUGAGUUAAUAAAAUU .(((((.................((((...........))))....((((((....))))))....))))).........((((((....)))))).((((....))))........... ( -25.40) >DroEre_CAF1 53581 120 - 1 CGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAUACUUAUCGAACUAUCAGCAAAGGCAAAGGCGAAAUCUUGUGCUCCUCAAAAUUGAGUUAAUAAAGUU .(((((.................((((...........)))).....(((((....))))).....)))))...((((((((.....)))).)))).((((....))))........... ( -20.50) >DroYak_CAF1 47819 120 - 1 CGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAAACUUAUCGAACUAUCAGCUCAGGCGAAGGCGAAAUCUUUCGCCCCUCGGAGUUGAGUUAAUAAAGUU (((((((.....)).........((((...........))))..)))))...(((((....(((..(((((((.((((((((......)))))))).....))))))))))....))))) ( -32.20) >consensus CGCUGACAAUUUGUACUCUUUAAUGGUGUUUAAUUAAAACCAAUCAGCGAUAAACUUAUCGAACUAUCAGCUAAGGCAAAGGCGAAAUCUUUCGCCCCUCAAAAUUGAGUUAAUAAAAUU .(((((.................((((...........)))).....(((((....))))).....))))).........((((((....)))))).((((....))))........... (-21.02 = -20.94 + -0.08)

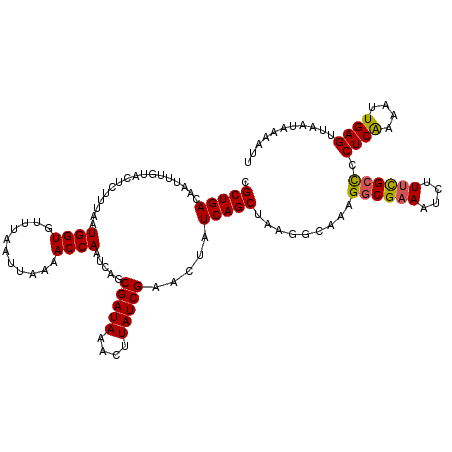
| Location | 379,007 – 379,127 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 379007 120 + 22407834 UUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCAGUGCAGUCAGCGCCAAUAAAUCACGUUUUGUCUAGCUGAU ......(((((((((((..((....(((((((((((((((((((.....)))).................((((....)))).)))))))))...))))))..))..))))).)))))). ( -29.90) >DroSec_CAF1 46645 120 + 1 UUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCGGUGCAGUCAGCGCCAAUAAAUCACGUUUUGUCUAGCUGAU ......(((((((((((..((....((((((.((((((((.(((((((........)))))))....))....))))))(((((.....))))).))))))..))..))))).)))))). ( -31.20) >DroSim_CAF1 47307 120 + 1 UUUGCCUUAGCUGAUAGUACGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCGGUGCAGUCAGCACCAAUAAAUCACGUUUUGUCUAGCUGAU ......(((((((((((.(((....((((((.((((((((.(((((((........)))))))....))....))))))(((((.....))))).))))))..))).))))).)))))). ( -32.10) >DroEre_CAF1 53621 120 + 1 UUUGCCUUUGCUGAUAGUUCGAUAAGUAUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCGGUGCAGUCAGCGCCAAUAAAUCACGUUUUGUCUAGCUGAU ..(((..(((((((((((((((((....)))))((...((((.((.....)).))))....))......)))))))))))).)))((((((....(((((......)))))...)))))) ( -30.40) >DroYak_CAF1 47859 120 + 1 UUCGCCUGAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCGGUGCAGUCAGCGCCAAUAAAUCACGUUUUGUCUAGCUGAU ........(((((((((..((....((((((.((((((((.(((((((........)))))))....))....))))))(((((.....))))).))))))..))..))))).))))... ( -29.30) >consensus UUUGCCUUAGCUGAUAGUUCGAUAAGUUUAUCGCUGAUUGGUUUUAAUUAAACACCAUUAAAGAGUACAAAUUGUCAGCGGUGCAGUCAGCGCCAAUAAAUCACGUUUUGUCUAGCUGAU ......(((((((((((..((...........((((((((.(((((((........)))))))....))....))))))(((((.....))))).........))..))))).)))))). (-28.00 = -28.44 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:25 2006