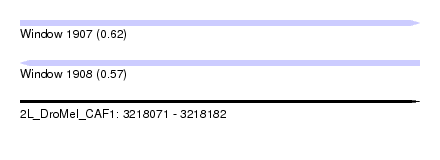
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,218,071 – 3,218,182 |
| Length | 111 |
| Max. P | 0.615304 |

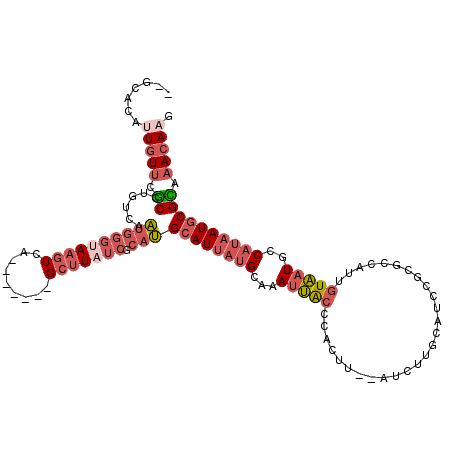
| Location | 3,218,071 – 3,218,182 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
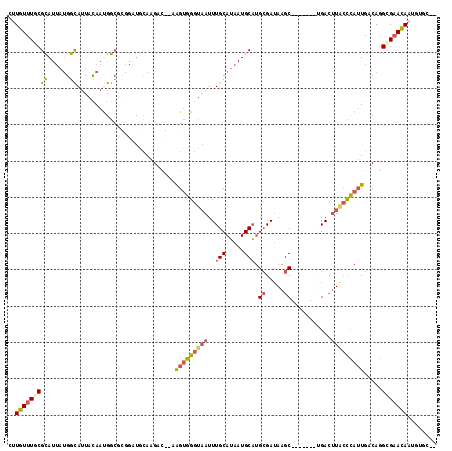
>2L_DroMel_CAF1 3218071 111 + 22407834 CUUGUUUGCGCAUUAUGGCAUUACAAUGGCGCGGAUGCAAGAUGCAAGUGGGCAAUUUGCAUAAUGCAUGCGAUAAGC-------UGACUUACCCAUUGACAGGCGAAUAAUGUGCAU ......((((((((((.((....((((((..((.(((((..((((((((.....))))))))..))))).)).((((.-------...)))).))))))....))..)))))))))). ( -41.60) >DroSec_CAF1 52160 118 + 1 CUUGUUUGCGCAUUAUGGCAUUACAAUGGCACGGAUACAAGAUGCAAGUGGGUAAUUUGCAUAAUGCAUGCGAUAAGCUUAUCGCUGACUUACCCAUUGACAGGCGAACAAUGUGCAU .(((((((((((((.((.(((....))).)).(....)..))))).(((((((((..(((.....))).((((((....))))))....))))))))).....))))))))....... ( -36.20) >DroSim_CAF1 50640 111 + 1 CUUGUUUGCGCAUUAUGGCAUUACAAUGGCGCGGAUACAAGAUGCAAGUGGGUAAUUUGCAUAAUGCAUGCGAUAAGC-------UGACUUACCCAUUGACAGGCGAACAAUGUGCAU ......((((((((.(.((....((((((((((....((..((((((((.....))))))))..))..)))).((((.-------...)))).))))))....)).)..)))))))). ( -32.70) >DroEre_CAF1 48769 102 + 1 CUUGUUUACGCAUUAUGGCAUUACAACGGCGCGGAUGCAAGAC---AGUGGGUAAUUUGCAUAAUGCAUGCGAUAAGC-------UGACUUGCCCAUUGACAGGCGAACAAU------ .(((((..((((((...((.........))...)))))....(---(((((((((..(((.....))).((.....))-------....))))))))))....)..))))).------ ( -28.60) >DroYak_CAF1 47393 98 + 1 AAUGUUUACGCAUUAUGGCAUUACAACGG----GUUGCAAGAC---AGUGGGUAAUUUGCAUAAUGCAUGCGAAAAGC-------CGACUUGCUUAUUGACAAGCGAACAAU------ ..((((..(((((((((..(((((.((..----(((....)))---.))..)))))...)))))))).(((((.((((-------......)))).))).)).)..))))..------ ( -24.90) >DroPer_CAF1 62016 104 + 1 GGUGUUUUCGUAU---GGUAGAAUAAUAGCA------UAAGAC--CGGGGGCUCAUUUGCAAAAUGCUGGGGAUACGCUGAUAGGAGAACUA-CCAAUGACAGGAGCACAAUGUAC-- .((((((..((.(---(((((..((.((((.------((...(--(((.(((......))....).))))...)).)))).))......)))-)))...))..)))))).......-- ( -25.50) >consensus CUUGUUUGCGCAUUAUGGCAUUACAAUGGCGCGGAUGCAAGAC__AAGUGGGUAAUUUGCAUAAUGCAUGCGAUAAGC_______UGACUUACCCAUUGACAGGCGAACAAUGUGC__ ..(((((((((......))...........................(((((((((..(((.....))).((.....))...........))))))))).....)))))))........ (-14.50 = -15.17 + 0.67)

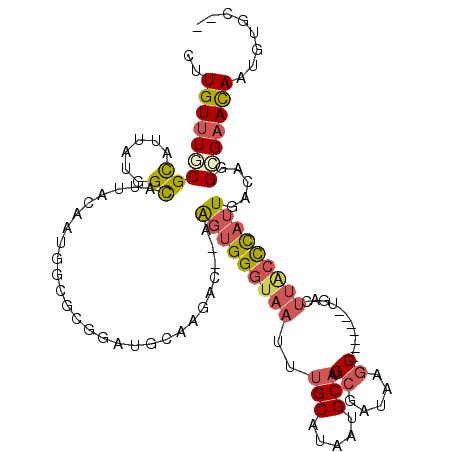
| Location | 3,218,071 – 3,218,182 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -11.73 |
| Energy contribution | -13.60 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3218071 111 - 22407834 AUGCACAUUAUUCGCCUGUCAAUGGGUAAGUCA-------GCUUAUCGCAUGCAUUAUGCAAAUUGCCCACUUGCAUCUUGCAUCCGCGCCAUUGUAAUGCCAUAAUGCGCAAACAAG .(((.((((((..((....(((((((((((...-------.)))))((((((((..((((((.........))))))..)))))..)))))))))....)).)))))).)))...... ( -34.80) >DroSec_CAF1 52160 118 - 1 AUGCACAUUGUUCGCCUGUCAAUGGGUAAGUCAGCGAUAAGCUUAUCGCAUGCAUUAUGCAAAUUACCCACUUGCAUCUUGUAUCCGUGCCAUUGUAAUGCCAUAAUGCGCAAACAAG .(((.((((((..((....((((((........((((((....))))))(((((..((((((.........))))))..))))).....))))))....)).)))))).)))...... ( -34.60) >DroSim_CAF1 50640 111 - 1 AUGCACAUUGUUCGCCUGUCAAUGGGUAAGUCA-------GCUUAUCGCAUGCAUUAUGCAAAUUACCCACUUGCAUCUUGUAUCCGCGCCAUUGUAAUGCCAUAAUGCGCAAACAAG .(((.((((((..((....(((((((((((...-------.)))))((((((((..((((((.........))))))..)))))..)))))))))....)).)))))).)))...... ( -32.80) >DroEre_CAF1 48769 102 - 1 ------AUUGUUCGCCUGUCAAUGGGCAAGUCA-------GCUUAUCGCAUGCAUUAUGCAAAUUACCCACU---GUCUUGCAUCCGCGCCGUUGUAAUGCCAUAAUGCGUAAACAAG ------.(((((.((((......))))......-------((.....))((((((((((...(((((..((.---((...((....)))).)).)))))..)))))))))).))))). ( -24.60) >DroYak_CAF1 47393 98 - 1 ------AUUGUUCGCUUGUCAAUAAGCAAGUCG-------GCUUUUCGCAUGCAUUAUGCAAAUUACCCACU---GUCUUGCAAC----CCGUUGUAAUGCCAUAAUGCGUAAACAUU ------...((..((((((......))))))..-------)).......((((((((((.............---((.((((((.----...)))))).))))))))))))....... ( -21.81) >DroPer_CAF1 62016 104 - 1 --GUACAUUGUGCUCCUGUCAUUGG-UAGUUCUCCUAUCAGCGUAUCCCCAGCAUUUUGCAAAUGAGCCCCCG--GUCUUA------UGCUAUUAUUCUACC---AUACGAAAACACC --.......(((....(((...(((-(((......((..((((((...((.((.....))....(......))--)...))------))))..))..)))))---).)))....))). ( -16.20) >consensus __GCACAUUGUUCGCCUGUCAAUGGGUAAGUCA_______GCUUAUCGCAUGCAUUAUGCAAAUUACCCACUU__AUCUUGCAUCCGCGCCAUUGUAAUGCCAUAAUGCGCAAACAAG .......(((((.((......((((((((((.........))))))).)))((((((((...(((((...........................)))))..)))))))))).))))). (-11.73 = -13.60 + 1.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:32 2006