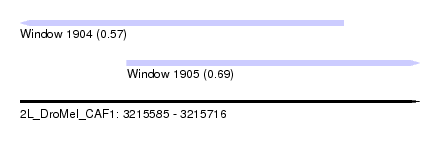
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,215,585 – 3,215,716 |
| Length | 131 |
| Max. P | 0.693779 |

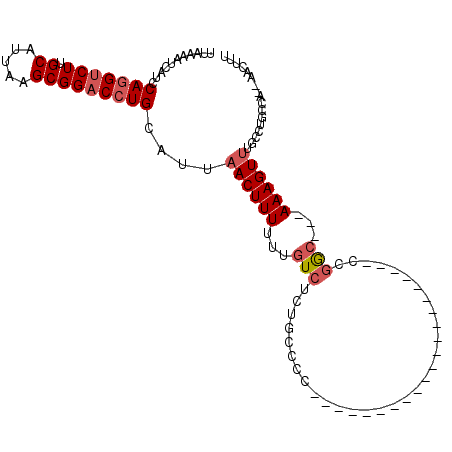
| Location | 3,215,585 – 3,215,691 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -21.37 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3215585 106 - 22407834 UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUCUUUGCCCCCGCAGCCCCCCUGCAACCCAUCCGGC---AAAGUUUGCCUGCCA--AACUUU ...........(((((((.((.....)))))))))................((((((...((((.....)))).........)))---)))(((((.....))--)))... ( -22.40) >DroSec_CAF1 49721 85 - 1 UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUCUUUGCCCC---------------------CCGGC---AAAGUUUGCCUGCCA--AACUUU ...........(((((((.((.....)))))))))................((((((..---------------------..)))---)))(((((.....))--)))... ( -20.70) >DroSim_CAF1 48163 85 - 1 UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUCUUUGCCCC---------------------CCGGC---AAAGUUUGCCUGCCA--AACUUU ...........(((((((.((.....)))))))))................((((((..---------------------..)))---)))(((((.....))--)))... ( -20.70) >DroEre_CAF1 46345 90 - 1 UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUCUCUGCCCCCCCGG----------------CCGGC---AAAGUUUGCCUGCCA--AACUUU ...........(((((((.((.....)))))))))...........................((----------------(.(((---(.....)))).))).--...... ( -23.60) >DroYak_CAF1 44991 93 - 1 UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUUUCUGCCCCCCAAA----------------CCCGGAAAAAAGUUUGCCUGCCA--AACUUU .........(((((((((.((.....))))))))(((..(((......)))..)))........----------------...)))..((((((((.....))--)))))) ( -22.50) >DroPer_CAF1 59117 86 - 1 UUAAAAUCACCCAAGACUGGCGUUAAGCGGACCUGCAUU-ACUUUUUUGUCUCUGUCAA---------------------ACGAC---AAAGUUGAAGCGCCAAAAACUUU .................(((((((..(((....)))...-.(..(((((((........---------------------..)))---))))..).)))))))........ ( -18.30) >consensus UUAAAAUCAUCCAGGUCUUGCAUUAAGCGGACCUGCAUUAACUUUUUUGUCUCUGCCCC_____________________CCGGC___AAAGUUUGCCUGCCA__AACUUU ...........(((((((.((.....)))))))))....((((((...(((...............................)))...))))))................. (-10.95 = -11.65 + 0.69)

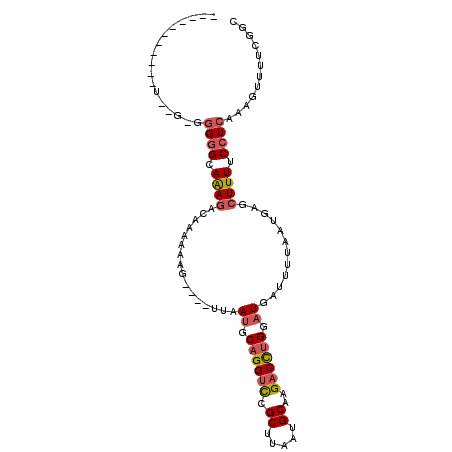
| Location | 3,215,620 – 3,215,716 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -13.81 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3215620 96 + 22407834 AGGGG---GGCU--GCGGGGGCAAAGACAAAAAAG----UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUCGGC ..(..---((((--...((((.((((.((..((((----(((.(.((((((.((.....))..)))))).))))))))..))..)))).))))..))))..)... ( -30.90) >DroSec_CAF1 49747 84 + 1 -----------------GGGGCAAAGACAAAAAAG----UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUCGGC -----------------((((.((((.((..((((----(((.(.((((((.((.....))..)))))).))))))))..))..)))).))))............ ( -23.10) >DroSim_CAF1 48189 84 + 1 -----------------GGGGCAAAGACAAAAAAG----UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUCGGC -----------------((((.((((.((..((((----(((.(.((((((.((.....))..)))))).))))))))..))..)))).))))............ ( -23.10) >DroEre_CAF1 46371 89 + 1 ----------CC--GGGGGGGCAGAGACAAAAAAG----UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUUGGC ----------((--....)).(((((((...((((----(((.(.((((((.((.....))..)))))).))))))))..((((......))))..))))))).. ( -24.50) >DroWil_CAF1 53513 101 + 1 AAGAAAAACGAUGAGAAAGAGCAAAGAAGUAAAAGAAUCUUAAUGCAGGUUCGCUUAGUGCAAGUCUGGGCUUAUUUUAAUGAGCUUUUCGUCGACUAUUU---- ........(((((((((.(..((..(((((((..((((((......))))))((((((.......)))))))))))))..))..)))))))))).......---- ( -26.60) >DroYak_CAF1 45020 89 + 1 ----------UU--UGGGGGGCAGAAACAAAAAAG----UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUUGGC ----------((--(((((((.((...((..((((----(((.(.((((((.((.....))..)))))).))))))))..))..)).)))))))))......... ( -25.60) >consensus ___________U__G_GGGGGCAAAGACAAAAAAG____UUAAUGCAGGUCCGCUUAAUGCAAGACCUGGAUGAUUUUAAUGAGCUUUUCCUCAAAGUUUUCGGC .................((((.((((................((.((((((.((.....))..)))))).))............)))).))))............ (-13.81 = -14.31 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:29 2006