
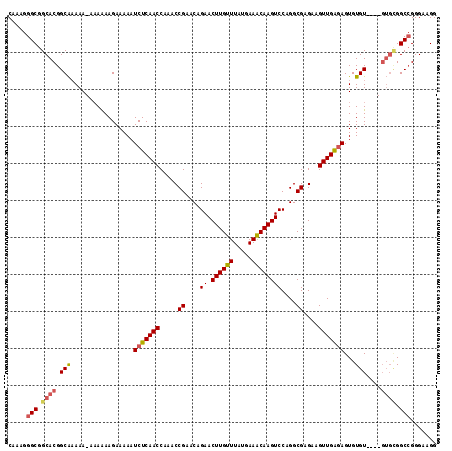
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,213,776 – 3,213,925 |
| Length | 149 |
| Max. P | 0.949115 |

| Location | 3,213,776 – 3,213,887 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
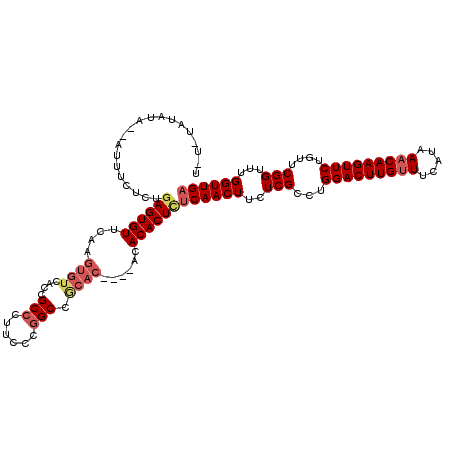
>2L_DroMel_CAF1 3213776 111 + 22407834 CAAAGGGCGGCACGGCAAAAA-AAAAAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAACAAGUCCAGGCGAGAAGUUGGGAGUGUGU----GUGCGGCCGGGAAGG .....(((.((((((((....-............(((((((.....((....((.((((((.....))))))))....))....))))))).))).)----)))).)))....... ( -28.13) >DroSec_CAF1 47917 110 + 1 CAAAGGGCGGCACGGCAAAAA--AAAAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAGCAAGUCCAGGCGAGAAGUUGAGAGUGUGU----GUGCGGCAGGGAAGG ......((.((((((((....--...........(((((((.....((....((.((((((.....))))))))....))....))))))).))).)----)))).))........ ( -25.79) >DroSim_CAF1 46346 109 + 1 CAAAGGGCGGCACGGCAAAAA---AAAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAACAAGUCCAGGCGAGAAGUUGAGAGUGUGU----GUGCGGCCGGGAAGG .....(((.((((((((....---..........(((((((.....((....((.((((((.....))))))))....))....))))))).))).)----)))).)))....... ( -29.16) >DroEre_CAF1 44502 109 + 1 CAAAGGGCGGCACGGCGCAAA-AAAGAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAACAAGUCCAGGCGAGAAGUUGAAAGUGUGUGC------GGCCGGGAAGG .....(((.(((((.(((...-......((....))(((((.....((....((.((((((.....))))))))....))....)))))..))))))))------.)))....... ( -27.30) >DroYak_CAF1 43093 116 + 1 CAAAGGGCGGCACGGCAAAAAAAAAAAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAACAAGUCCAGGCGAGCAGUUGAGAGUGUGUGCGUGUGUGGCCGGGAAGG .....(((.((((((((.................(((((((.....((....((.((((((.....))))))))....))....))))))).....))).))))).)))....... ( -29.95) >consensus CAAAGGGCGGCACGGCAAAAA_AAAAAAGAAAAAUCUCAACCAAACCGAACAGAACUUGUUUAUGAAACAAGUCCAGGCGAGAAGUUGAGAGUGUGU____GUGCGGCCGGGAAGG .....(((.((((.(((.................(((((((.....((....((.((((((.....))))))))....))....)))))))...)))....)))).)))....... (-21.55 = -22.15 + 0.60)


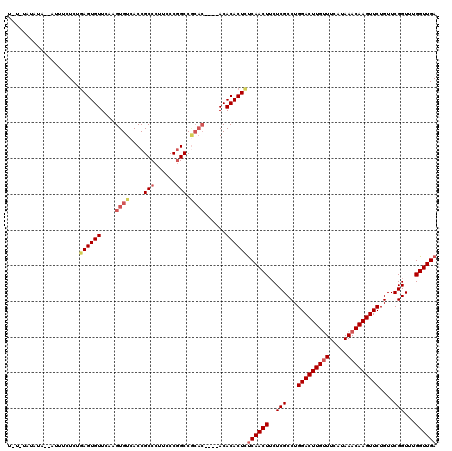
| Location | 3,213,776 – 3,213,887 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -20.76 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3213776 111 - 22407834 CCUUCCCGGCCGCAC----ACACACUCCCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUUUUU-UUUUUGCCGUGCCGCCCUUUG .......(((.((((----.........(((((..(((...(((((((((.....)))))))))....)))...)))))(((....))).....-........)))).)))..... ( -28.00) >DroSec_CAF1 47917 110 - 1 CCUUCCCUGCCGCAC----ACACACUCUCAACUUCUCGCCUGGACUUGCUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUUUU--UUUUUGCCGUGCCGCCCUUUG ........((.((((----......((((((((....(((.((((..((((.......))))...)))))))..)))))))).....(.....--.....)..)))).))...... ( -23.10) >DroSim_CAF1 46346 109 - 1 CCUUCCCGGCCGCAC----ACACACUCUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUUU---UUUUUGCCGUGCCGCCCUUUG .......(((.((((----......((((((((..(((...(((((((((.....)))))))))....)))...)))))))).....(....---.....)..)))).)))..... ( -29.80) >DroEre_CAF1 44502 109 - 1 CCUUCCCGGCC------GCACACACUUUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUCUUU-UUUGCGCCGUGCCGCCCUUUG .......(((.------((((......((((((..(((...(((((((((.....)))))))))....)))...))))))((....))......-........)))).)))..... ( -28.80) >DroYak_CAF1 43093 116 - 1 CCUUCCCGGCCACACACGCACACACUCUCAACUGCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUUUUUUUUUUUGCCGUGCCGCCCUUUG .......(((..(((..(((.....(((((((((((.((..(((((((((.....))))))))).))..)))..)))))))).................))).)))..)))..... ( -27.05) >consensus CCUUCCCGGCCGCAC____ACACACUCUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGAGAUUUUUCUUUUUU_UUUUUGCCGUGCCGCCCUUUG .......(((.((((..........((((((((..(((...(((((((((.....)))))))))....)))...)))))))).....(............)..)))).)))..... (-20.76 = -22.20 + 1.44)


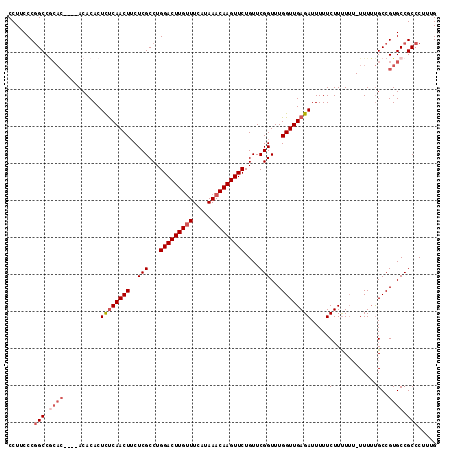
| Location | 3,213,811 – 3,213,925 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3213811 114 - 22407834 UUUUUAUAUA--AUUUUUCUGAGUGUUCAAGUGUCACCGCCCUUCCCGGCCGCAC----ACACACUCCCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA ..........--........((((((....((((..(((.......)))..))))----..)))))).(((((..(((...(((((((((.....)))))))))....)))...))))). ( -29.00) >DroSec_CAF1 47951 114 - 1 UCUCUCUAUA--AUUUCUCUGAGUGUUCAAGUGUCACCGCCCUUCCCUGCCGCAC----ACACACUCUCAACUUCUCGCCUGGACUUGCUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA ..........--........((((((....((((....((........)).))))----..))))))((((((....(((.((((..((((.......))))...)))))))..)))))) ( -23.00) >DroSim_CAF1 46379 114 - 1 UCUCUCUAUA--AUUUUUCUGAGUGUUGAAGUGUCACCGCCCUUCCCGGCCGCAC----ACACACUCUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA ..........--........((((((....((((..(((.......)))..))))----..))))))((((((..(((...(((((((((.....)))))))))....)))...)))))) ( -30.00) >DroEre_CAF1 44537 110 - 1 ----UAUAUAAUAUUUCUCUGAGUGUUCUAGUGUCACCGCCCUUCCCGGCC------GCACACACUUUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA ----................((((((....((((..(((.......)))..------))))))))))((((((..(((...(((((((((.....)))))))))....)))...)))))) ( -27.20) >DroYak_CAF1 43129 106 - 1 --------------UUCUCUCAGUGUUGAAGUGUCACCGCCCUUCCCGGCCACACACGCACACACUCUCAACUGCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA --------------.......(((((....((((....(((......)))...))))....))))).(((((((((.((..(((((((((.....))))))))).))..)))..)))))) ( -27.70) >consensus U_U_UAUAUA__AUUUCUCUGAGUGUUCAAGUGUCACCGCCCUUCCCGGCCGCAC____ACACACUCUCAACUUCUCGCCUGGACUUGUUUCAUAAACAAGUUCUGUUCGGUUUGGUUGA ....................((((((....((((....(((......))).))))......))))))((((((..(((...(((((((((.....)))))))))....)))...)))))) (-22.62 = -23.98 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:26 2006