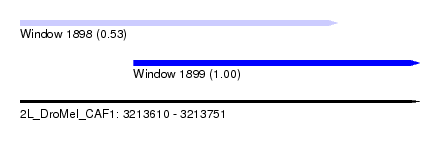
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,213,610 – 3,213,751 |
| Length | 141 |
| Max. P | 0.999765 |

| Location | 3,213,610 – 3,213,722 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
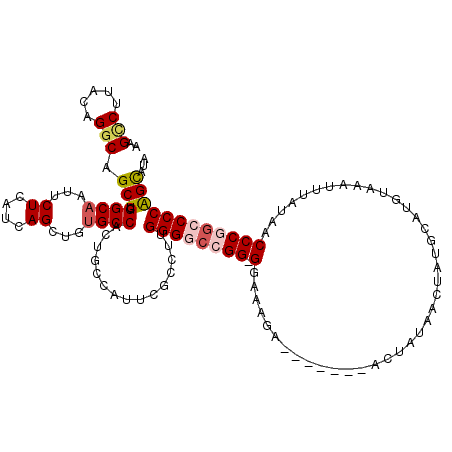
>2L_DroMel_CAF1 3213610 112 + 22407834 AAGCCUUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACUGCCAUUCGCCUUGGGGCCGG-GGAAAGA-------ACUAUAACUAUGCAUGUAAAUUUAUAACCCGACCCCGGCAUU ..(((.....(((((...((((...((....))...)))).))))).........((((.(((-(...(((-------..((((........))))..)))....)))).)))))))... ( -35.00) >DroPse_CAF1 61034 108 + 1 AAGUCGUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACU------------GGGGCAGGGGGAGAGAAAUCACUACUAUAACUAUUCCAGUAAAUUUAUAACCCCGACCCAGUCGA ...(((....((((...((((...........))))))))(((------------(((...((((..((....))..((((...........)))).........))))..))))))))) ( -30.80) >DroSim_CAF1 46186 112 + 1 AAGCCUUACAGGCAGCUCGGCAAUUCUCAUCAGCUGCGCCACUGCCAUUCGCCUUGGGGCCGG-GGAAAGA-------ACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCGGCAUU ..(((.....((((((.((((...........)))).))...)))).........((((((((-(...(((-------..((((........))))..)))....))))))))))))... ( -40.40) >DroEre_CAF1 44337 112 + 1 AAGGCUUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACUGCCAUUCGCCUUGGGGCCGG-GGAAAGA-------ACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCAGCAUC (((((.....(((((...((((...((....))...)))).)))))....)))))((((((((-(...(((-------..((((........))))..)))....)))))))))...... ( -41.70) >DroYak_CAF1 42945 112 + 1 AAGCCUUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACUGCCAUUCGCCUUGGGGCCGG-GGAAAGA-------ACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCAGUAUA ......(((.(((((...((((...((....))...)))).)))))........(((((((((-(...(((-------..((((........))))..)))....))))))))))))).. ( -38.60) >DroPer_CAF1 57066 108 + 1 AAGCCGUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACU------------GGGGCAGGGGGAGAGAAAUCACUACUAUAACUAUUCAAGUAAAUUUAUAACCCCGACCCAGUCGA ..(((.....))).((.((((...........)))).)).(((------------(((...((((..((....))..((((...........)))).........))))..))))))... ( -33.30) >consensus AAGCCUUACAGGCAGCUCGGCAAUUCUCAUCAGCUGUGCCACUGCCAUUCGCCUUGGGGCCGG_GGAAAGA_______ACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCAGCAUA ..(((.....))).(((.((((...((....))...))))...............((((((((.(........................................))))))))))))... (-21.95 = -22.53 + 0.58)

| Location | 3,213,650 – 3,213,751 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -23.16 |
| Energy contribution | -22.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
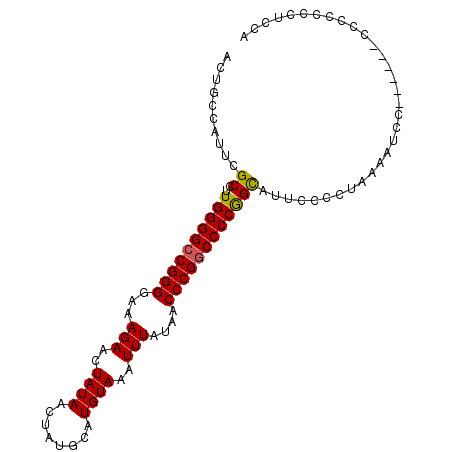
>2L_DroMel_CAF1 3213650 101 + 22407834 ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGACCCCGGCAUUCCCCUAAAAUCCAACCCCCCCCCCCUCCA ..........(((..((((.((((...(((..((((........))))..)))....)))).)))))))................................ ( -19.80) >DroSec_CAF1 47797 95 + 1 ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCGGCAUUCCCCUAAAAUCC------CGCCCCCUCCA ..........(((..(((((((((...(((..((((........))))..)))....))))))))))))...............------........... ( -26.90) >DroSim_CAF1 46226 95 + 1 ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCGGCAUUUCCCUAAAAUCC------CGACCCCUCCA ..........(((..(((((((((...(((..((((........))))..)))....))))))))))))...............------........... ( -26.90) >DroEre_CAF1 44377 100 + 1 ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCAGCAUCCCCCUAAAAUCCCAGCC-CCCUCCCUCCA ..........((..((((((((((...(((..((((........))))..)))....))))))))))))....................-........... ( -25.20) >DroYak_CAF1 42985 85 + 1 ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCAGUAUACCCCUAAAAUCCC---------------- .............(((((((((((...(((..((((........))))..)))....))))))))))).................---------------- ( -23.90) >consensus ACUGCCAUUCGCCUUGGGGCCGGGGAAAGAACUAUAACUAUGCAUGUAAAUUUAUAACCCGGCCCCGGCAUUCCCCUAAAAUCC______CCCCCCCUCCA ..........((..((((((((((...(((..((((........))))..)))....))))))))))))................................ (-23.16 = -22.96 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:24 2006