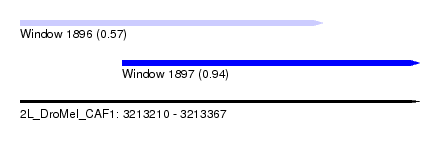
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,213,210 – 3,213,367 |
| Length | 157 |
| Max. P | 0.935974 |

| Location | 3,213,210 – 3,213,329 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3213210 119 + 22407834 AUCAGCGUGACGAAGUGAGUCCUGCAGCUUAAUGGAGAUAGAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAGCGGAAAUGAACCCAACGGAACUUUUUCCGCUGACCGAAAUGACA .((((((.((.(((((..((.((((.(((((((.((.((...)).)).)))))))...)))).))...((....))((........))......))))).))))))))........... ( -34.70) >DroSec_CAF1 47367 112 + 1 AUCAGCGUGAAGCAAUGAGUCC-------UAAUGGAGAUAGAAUGUCAAUUAAGUGACGCAGCAUCAAGCGGAAACGGAAACGAACCCAACGGAACCUUUUCCGCUGACCGAAAUGACA .(((.(((((.((......(((-------....)))........((((......))))...)).)))((((((((((....))..((....)).....))))))))...))...))).. ( -28.30) >DroSim_CAF1 45796 112 + 1 AUCAGCGUGACGAAGUGAGUCC-------UAAUGGAGAUAAAAUGUCAAUUAAGUGACGCAGCAUGAAGCGGAAACGGAAACGAACCCAGCGGAACCUUUUCCGCUGACCGAAAUGACA ....((((.((........(((-------....)))(((.....)))......)).)))).((((....(((...((....))....((((((((....)))))))).)))..))).). ( -35.30) >DroEre_CAF1 43940 119 + 1 AUCAGCGUGACGAAGUGAUUCCUGUAGCCUAAUGGAGAUAGAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAGCGGAAAUGAACCCAACGGAACCUUUUCCGCUGACCAAAAUGACA .(((((((.((....((((..((((..((....))..))))...)))).....)).)))).((.....))((.((((((((....((....)).....))))))))..))....))).. ( -35.40) >DroYak_CAF1 42547 119 + 1 AUCAGCGUGACGAAGUGAGUCGUGUUGCCUAAUGGAGAUAGAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAACGGAAAUGAACCCAACGAAACCCUUUCCGCUGACCGAAAUGACA .(((((........((...((((((((((((.......)))...((((......))))))))))))).))(((((.((.................)).))))))))))........... ( -32.63) >consensus AUCAGCGUGACGAAGUGAGUCCUG__GC_UAAUGGAGAUAGAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAACGGAAAUGAACCCAACGGAACCUUUUCCGCUGACCGAAAUGACA .(((((((.((........(((...........)))(((.....)))......)).)))).......((((((((.((...((.......))...)).))))))))........))).. (-23.38 = -23.14 + -0.24)

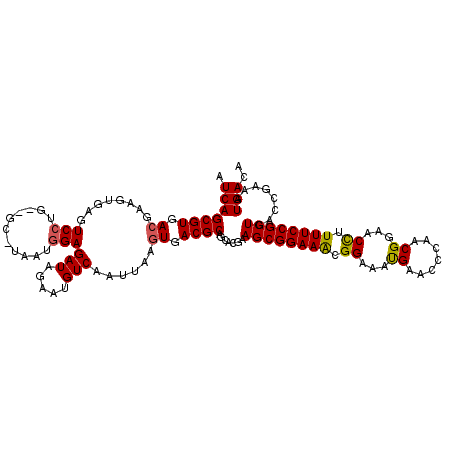
| Location | 3,213,250 – 3,213,367 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -26.94 |
| Energy contribution | -26.02 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3213250 117 + 22407834 GAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAGCGGAAAUGAACCCAACGGAACUUUUUCCGCUGACCGAAAUGACAAAUGCUUGUCAUAAAGCGCCUUGGCCUAGCUUUAAGCU ....((((......))))..(((..(((((((.((((((((....((....)).....))))))))..))...(((((((....)))))))..((.((....)))).)))))..))) ( -34.80) >DroSec_CAF1 47400 117 + 1 GAAUGUCAAUUAAGUGACGCAGCAUCAAGCGGAAACGGAAACGAACCCAACGGAACCUUUUCCGCUGACCGAAAUGACAAAUGCUUGUCAUAAAGCGCCUUGGUCUAGCUUUAAGCU ....((((......))))(((((....((((((((((....))..((....)).....))))))))((((((.(((((((....)))))))........))))))..)))....)). ( -33.10) >DroSim_CAF1 45829 117 + 1 AAAUGUCAAUUAAGUGACGCAGCAUGAAGCGGAAACGGAAACGAACCCAGCGGAACCUUUUCCGCUGACCGAAAUGACAAAUGCAUGUCAUAAAGCGCCUUGGCCUAGCUUUAAGCU ....((((......))))..(((.((((((((...((....))....((((((((....)))))))).))...((((((......))))))..((.((....)))).)))))).))) ( -39.90) >DroEre_CAF1 43980 117 + 1 GAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAGCGGAAAUGAACCCAACGGAACCUUUUCCGCUGACCAAAAUGACAAAUGCUUGUCGUAAAGCGCUUUUGACUAGCUUAAAGCU ....((((......))))((((((.(((((((.((((((((....((....)).....))))))))..))...(((((((....))))))).....))))))).)).))........ ( -32.60) >DroYak_CAF1 42587 117 + 1 GAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAACGGAAAUGAACCCAACGAAACCCUUUCCGCUGACCGAAAUGACAAAUGCGUGUCAUAAAGCGCUUUGGACUCGGUUUAAGCG ....((((......))))((.......((((((((.((.................)).))))))))((((((.....((((.((((........))))))))...))))))...)). ( -32.33) >consensus GAAUGUCAAUUAAGUGACGCAGCACGAAGCGGAAACGGAAAUGAACCCAACGGAACCUUUUCCGCUGACCGAAAUGACAAAUGCUUGUCAUAAAGCGCCUUGGACUAGCUUUAAGCU ....((((......))))((.((....((((((((.((...((.......))...)).)))))))).......((((((......))))))...))))........(((.....))) (-26.94 = -26.02 + -0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:22 2006