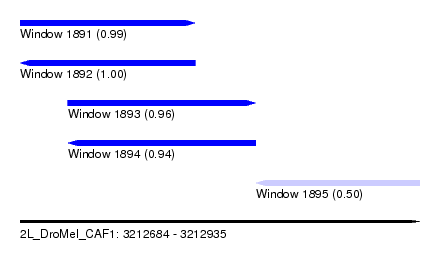
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,212,684 – 3,212,935 |
| Length | 251 |
| Max. P | 0.999763 |

| Location | 3,212,684 – 3,212,794 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3212684 110 + 22407834 G---UUCCACCCAUUUCUGGGCAAAGUUAAUGUCACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAU .---.....((((....))))........((((.(((((...((((.(((((((.((.((((.((.......))..)))).))....))))))).))))..)))))...)))) ( -30.60) >DroSec_CAF1 46835 105 + 1 --------GCCCAUCUCUGGGCAAAGUUAAUGUCACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAU --------(((((....))))).......((((.(((((...((((.(((((((.((.((((.((.......))..)))).))....))))))).))))..)))))...)))) ( -34.80) >DroSim_CAF1 45254 110 + 1 G---CUUCGCCCAUCUCUGGGCAAAGUUAAUGUCACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAU (---(((.(((((....))))).))))..((((.(((((...((((.(((((((.((.((((.((.......))..)))).))....))))))).))))..)))))...)))) ( -37.10) >DroEre_CAF1 43439 110 + 1 G---CACCACCAGUCUCUAGGCAAAGUUAAUGUCACGCAUUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUAUUACAAGUCAGCUGGCUGCGUUUUACAU (---((...(((((.....(((...((.((((.....)))))).)))(((((((.((.((((.((.......))..)))).))....))))))).))))).)))......... ( -28.30) >DroYak_CAF1 41915 113 + 1 GCUGCACCACCAGACUCUAGGCAAAGUUAAUGUCACGCAUUGCAGCCAGCUUGUUGGCAUGACACUUUUAAUGUCCUCAUCGCUUUCACAAGUCAGCUGGCUGCGUUUUACAU ...........((((....((((.......)))).......((((((((((((..(((((((.((.......))..)))).)))..))......))))))))))))))..... ( -29.10) >consensus G___CACCACCCAUCUCUGGGCAAAGUUAAUGUCACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAU .........((((....))))........((((.(((((...((((.(((((((.((.((((.((.......))..)))).))....))))))).))))..)))))...)))) (-26.42 = -26.98 + 0.56)



| Location | 3,212,684 – 3,212,794 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -32.92 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3212684 110 - 22407834 AUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUGACAUUAACUUUGCCCAGAAAUGGGUGGAA---C ((((...(((((..((((((.(((((....((.((...(((((.....))))))).)).))))).))))))...))))).))))....((..((((....))))..)).---. ( -40.90) >DroSec_CAF1 46835 105 - 1 AUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUGACAUUAACUUUGCCCAGAGAUGGGC-------- ((((...(((((..((((((.(((((....((.((...(((((.....))))))).)).))))).))))))...))))).)))).......(((((....)))))-------- ( -36.70) >DroSim_CAF1 45254 110 - 1 AUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUGACAUUAACUUUGCCCAGAGAUGGGCGAAG---C ((((...(((((..((((((.(((((....((.((...(((((.....))))))).)).))))).))))))...))))).))))...(((((((((....)))))))))---. ( -43.30) >DroEre_CAF1 43439 110 - 1 AUGUAAAACGCAGCCAGCUGACUUGUAAUAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAAUGCGUGACAUUAACUUUGCCUAGAGACUGGUGGUG---C ((((...(((((..((((((.(((((....((.((...(((((.....))))))).)).))))).))))))...))))).)))).....(..((........))..)..---. ( -34.10) >DroYak_CAF1 41915 113 - 1 AUGUAAAACGCAGCCAGCUGACUUGUGAAAGCGAUGAGGACAUUAAAAGUGUCAUGCCAACAAGCUGGCUGCAAUGCGUGACAUUAACUUUGCCUAGAGUCUGGUGGUGCAGC .........((((((((((...((((....))))....(((((.....))))).........))))))))))..((((...(((((((((......)))).))))).)))).. ( -37.40) >consensus AUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUGACAUUAACUUUGCCCAGAGAUGGGUGGAG___C ((((...(((((..((((((.(((((....((.((...(((((.....))))))).)).))))).))))))...))))).)))).....(((((((....)))))))...... (-32.92 = -33.80 + 0.88)


| Location | 3,212,714 – 3,212,832 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 98.14 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.84 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3212714 118 + 22407834 CACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .....((((((((((((((((.((.((((.((.......))..)))).))....)))....))))))))))(((((((((((((.....)).))...)))))))))........))). ( -32.80) >DroSec_CAF1 46860 118 + 1 CACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .....((((((((((((((((.((.((((.((.......))..)))).))....)))....))))))))))(((((((((((((.....)).))...)))))))))........))). ( -32.80) >DroSim_CAF1 45284 118 + 1 CACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .....((((((((((((((((.((.((((.((.......))..)))).))....)))....))))))))))(((((((((((((.....)).))...)))))))))........))). ( -32.80) >DroEre_CAF1 43469 118 + 1 CACGCAUUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUAUUACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .((.(((((((((((((((((.((.((((.((.......))..)))).))....)))....))))))))))(((((((((((((.....)).))...)))))))))....)))).)). ( -34.80) >DroYak_CAF1 41948 118 + 1 CACGCAUUGCAGCCAGCUUGUUGGCAUGACACUUUUAAUGUCCUCAUCGCUUUCACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .((.((((((((((((((((..(((((((.((.......))..)))).)))..))......))))))))))(((((((((((((.....)).))...)))))))))....)))).)). ( -36.30) >consensus CACGCACUGCAGCCGGCUUGUUGCCAUGACACUUUUAAUGUCCUCAUCGCUUUUACAAGUCAGCUGGCUGCGUUUUACAUGCGCUUACUGCAGUCAAGUGUAAAAUGGAAAAUGAGUU .((.((..(((((((((((((.((.((((.((.......))..)))).))....)))....))))))))))(((((((((((((.....)).))...)))))))))......)).)). (-31.80 = -31.84 + 0.04)

| Location | 3,212,714 – 3,212,832 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 98.14 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.66 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3212714 118 - 22407834 AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((.(((..((..(.......)..))(((((.....))))).((....))))).)))))))))).)). ( -32.40) >DroSec_CAF1 46860 118 - 1 AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((.(((..((..(.......)..))(((((.....))))).((....))))).)))))))))).)). ( -32.40) >DroSim_CAF1 45284 118 - 1 AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((.(((..((..(.......)..))(((((.....))))).((....))))).)))))))))).)). ( -32.40) >DroEre_CAF1 43469 118 - 1 AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUAAUAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAAUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((.((((........))))......(((((.....)))))..(((.....))))))))))))).)). ( -32.60) >DroYak_CAF1 41948 118 - 1 AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUGAAAGCGAUGAGGACAUUAAAAGUGUCAUGCCAACAAGCUGGCUGCAAUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((((((...((((....))))....(((((.....))))).........)))))))))))))).)). ( -36.90) >consensus AACUCAUUUUCCAUUUUACACUUGACUGCAGUAAGCGCAUGUAAAACGCAGCCAGCUGACUUGUAAAAGCGAUGAGGACAUUAAAAGUGUCAUGGCAACAAGCCGGCUGCAGUGCGUG .((.((((.....(((((((..((.((......))))..))))))).((((((.(((..((..(.......)..))(((((.....))))).((....))))).)))))))))).)). (-31.90 = -31.66 + -0.24)

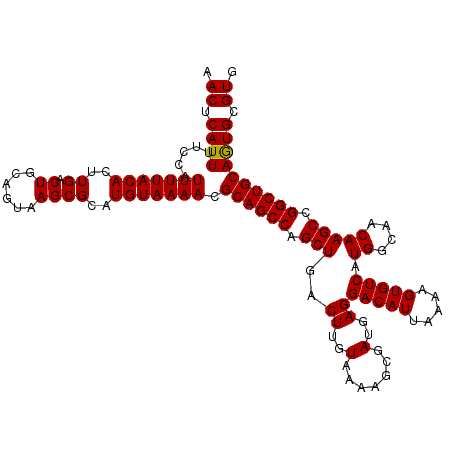
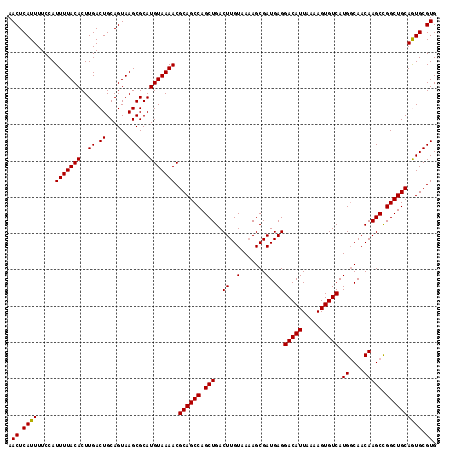
| Location | 3,212,832 – 3,212,935 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -24.28 |
| Energy contribution | -27.04 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3212832 103 - 22407834 GCCUGGCGAUAAGCCCGAGGCGAGUUGCCUUCGCCAACAAUCUUGGCUAAGUGCAGUUGCAUUCGGGCUCCUCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG ((((.(.((..((((((((((((.(((((((.(((((.....))))).))).)))))))).))))))))..)).)........((((....))))..)).)). ( -36.80) >DroSec_CAF1 46978 103 - 1 GCCUGGCGAUAAGCCCGAGGCGAGUUGCCUUCGCCAACAAUCUUGGCUAAGUGCAGUUGCAUUCGGGCUCCUCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG ((((.(.((..((((((((((((.(((((((.(((((.....))))).))).)))))))).))))))))..)).)........((((....))))..)).)). ( -36.80) >DroSim_CAF1 45402 103 - 1 GCCUGGCGAUAAGCCCGAGGCGAGUUGCCUUCGCCAACAAUCUUGGCUAAGUGCAGUUGCAUUCGGGCUCCUCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG ((((.(.((..((((((((((((.(((((((.(((((.....))))).))).)))))))).))))))))..)).)........((((....))))..)).)). ( -36.80) >DroEre_CAF1 43587 89 - 1 GCCUGGCGAUAAGCCCGAGGCGAGUUGCCAUCGCCAACAAUCUUGGCUAAGUG--------------CUCCGCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG (((((((.....)))..))))((((.(((...(((((.....)))))...).)--------------)...))))..((....((((....)))).....)). ( -22.90) >DroYak_CAF1 42066 103 - 1 GCCUGGCGAUAAGCUCGAGGCAAGUUGCCAUCGCCAACAAUCUUGGCUAAGUGCAGUUGCAUUCGGGCUCCGCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG ((((((((...((((((((((((.(((((...(((((.....)))))...).)))))))).)))))))).)))).........((((....))))..)).)). ( -32.40) >consensus GCCUGGCGAUAAGCCCGAGGCGAGUUGCCUUCGCCAACAAUCUUGGCUAAGUGCAGUUGCAUUCGGGCUCCUCUCAUGUUUACAGAUUUUAAUCUAAAGUGCG ((((...(((.((((((((((((.(((((((.(((((.....))))).))).)))))))).))))))))....((.((....)))).....)))...)).)). (-24.28 = -27.04 + 2.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:20 2006