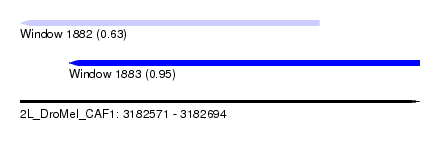
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,182,571 – 3,182,694 |
| Length | 123 |
| Max. P | 0.946057 |

| Location | 3,182,571 – 3,182,663 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -17.04 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
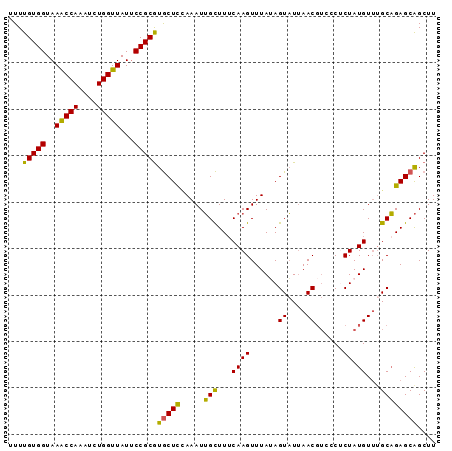
>2L_DroMel_CAF1 3182571 92 - 22407834 UUUUGUGGUAAGCCAAAUCUGGUUAUUCCGCGUGCUCUAAAUUGCUUGCAAGUUUAUAGUAUUAACGUCCCUCUAUGUUGGCAGAGCAGCUU ....((((..(((((....)))))...)))).((((((.....(((.(((((......((....))......)).))).))))))))).... ( -21.90) >DroSec_CAF1 19377 92 - 1 UUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUGUUUGCAGAGCAGCUU ....((((..(((((....)))))...))))..........(((((((((((..(((((.............))))))))).)))))))... ( -19.02) >DroSim_CAF1 18472 92 - 1 UUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUGUUUGCAGAGCAGCUU ....((((..(((((....)))))...))))..........(((((((((((..(((((.............))))))))).)))))))... ( -19.02) >DroEre_CAF1 19050 86 - 1 UUUUGUGGUAAACCAAAUUUGGUUAUUCCGCAUACUUUAAAUGGUUUUCAAGUUUAAAGUUU-AAC----CGCUAUGU-UGCCGAGCGGCUU ...(((((..(((((....)))))...))))).(((((((((.........)))))))))..-..(----((((.((.-...)))))))... ( -21.70) >DroYak_CAF1 18550 92 - 1 UUUUGUGGUAAACCAAAUUUGGUUAUGCCGCGUGCUCGAAAUUGCUUUCAAGUUUAAAGUAUUAACGUUUCACUAUGUUUGCAGAGCAGCUU ....(((((((((((....))))).)))))).(((((((((....)))).........(((..(((((......)))))))).))))).... ( -24.30) >consensus UUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUGUUUGCAGAGCAGCUU ...(((((..(((((....)))))...)))))(((((.....(((...((((......((....))......)).))...)))))))).... (-17.04 = -16.12 + -0.92)

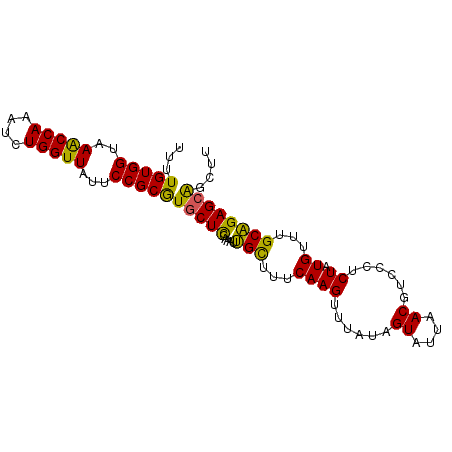
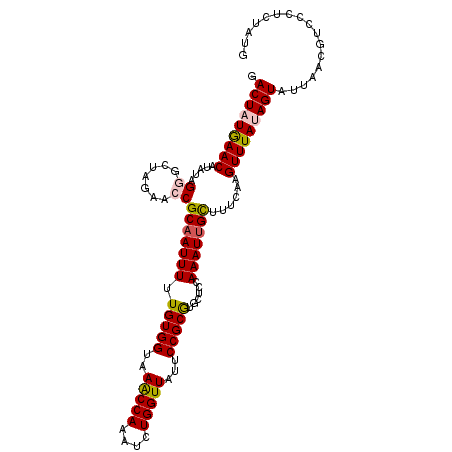
| Location | 3,182,586 – 3,182,694 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3182586 108 - 22407834 GACUAUAAACAUAUAGGGCUAGAACCGCAAUUUUUGUGGUAAGCCAAAUCUGGUUAUUCCGCGUGCUCUAAAUUGCUUGCAAGUUUAUAGUAUUAACGUCCCUCUAUG .(((((((((......((((...((((((.....)))))).))))......((.....))(((.((........)).)))..)))))))))................. ( -28.50) >DroSec_CAF1 19392 108 - 1 GACUAUGAACAUAUAGGGCUAGAACCGCAAUUUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUG .(((((((((.....((.......))(((((((.(((((..(((((....)))))...)))))......)))))))......)))))))))................. ( -26.80) >DroSim_CAF1 18487 108 - 1 GACUAUGAACAUAUAGGGCUAGAACCGCAAUUUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUG .(((((((((.....((.......))(((((((.(((((..(((((....)))))...)))))......)))))))......)))))))))................. ( -26.80) >DroEre_CAF1 19064 103 - 1 GACUGUGAACAUAUUGAAGUGGAACCGCAAUUUUUGUGGUAAACCAAAUUUGGUUAUUCCGCAUACUUUAAAUGGUUUUCAAGUUUAAAGUUU-AAC----CGCUAUG ((((.(((((...((((((((((((((((.....)))))).(((((....)))))..)))))..(((......))).)))))))))).)))).-...----....... ( -24.70) >DroYak_CAF1 18565 108 - 1 GACUGUGAACAUAUAGAACUGGAACCGCAAUUUUUGUGGUAAACCAAAUUUGGUUAUGCCGCGUGCUCGAAAUUGCUUUCAAGUUUAAAGUAUUAACGUUUCACUAUG ....((((((.....(((((.(((..((((((((((((((((((((....))))).))))))).....)))))))).))).)))))...((....))).))))).... ( -27.80) >consensus GACUAUGAACAUAUAGGGCUAGAACCGCAAUUUUUGUGGUAAACCAAAUCUGGUUAUUCCGCGUGCUCCAAAUUGCUUUCAAGUUUAUAGUAUUAACGUCCCUCUAUG .(((((((((.....((.......))(((((((.(((((..(((((....)))))...)))))......)))))))......)))))))))................. (-22.12 = -22.48 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:09 2006