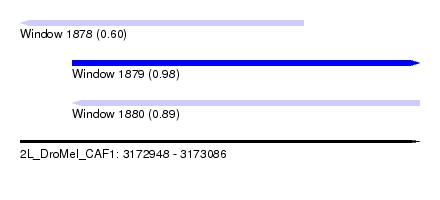
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,172,948 – 3,173,086 |
| Length | 138 |
| Max. P | 0.977396 |

| Location | 3,172,948 – 3,173,046 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3172948 98 - 22407834 UCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAGGUUUACGGAUUUUCUUUAUUUUUCAACAUUCUCUAUUCUAUUCUCUAUU .((((.....))))((((.(((((.(((........)))....(((((.(((.(..(((.......)))..).))).)))))...))))).))))... ( -17.90) >DroSec_CAF1 8512 95 - 1 UCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU-UACAAAUUU--UUUAUUUUUUAACAUUCUCGACUAUACUAAGCAUU ......((((...))))(((((((.......)))))......((((((.((-((.(((...--.....))).)))).))))))..........))... ( -22.40) >DroSim_CAF1 8959 91 - 1 UCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU-UACAAAUUU--UUUAUUUUUUAACAUUCUCGACUAUAC----CAUU .(((.(((((...))))).(((((.......)))))...)))((((((.((-((.(((...--.....))).)))).)))))).......----.... ( -21.90) >consensus UCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU_UACAAAUUU__UUUAUUUUUUAACAUUCUCGACUAUACU___CAUU .(((.(((((...))))).(((((.......)))))...)))((((((.((....((((.......))))....)).))))))............... (-17.37 = -17.27 + -0.11)

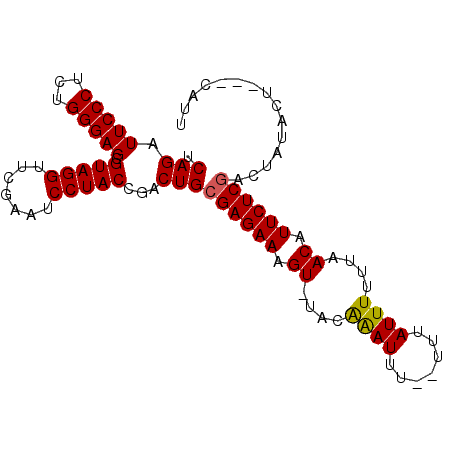
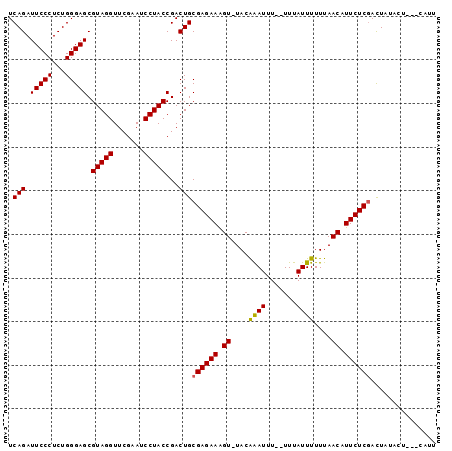
| Location | 3,172,966 – 3,173,086 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.30 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3172966 120 + 22407834 GAAUGUUGAAAAAUAAAGAAAAUCCGUAAACCUUCUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUG ...((((....))))......................((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).. ( -33.22) >DroSec_CAF1 8530 117 + 1 GAAUGUUAAAAAAUAAA--AAAUUUGUA-ACUUUCUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUG (((.((((.(((.....--...))).))-)).)))..((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).. ( -34.62) >DroSim_CAF1 8973 117 + 1 GAAUGUUAAAAAAUAAA--AAAUUUGUA-ACUUUCUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUG (((.((((.(((.....--...))).))-)).)))..((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).. ( -34.62) >DroYak_CAF1 8710 109 + 1 AAGU---AAA-----AA--AGAAUUUUA-CUUCUCUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUG ((((---(((-----(.--....)))))-))).....((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).. ( -35.52) >consensus GAAUGUUAAAAAAUAAA__AAAUUUGUA_ACUUUCUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUG .....................................((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).. (-32.02 = -32.02 + -0.00)

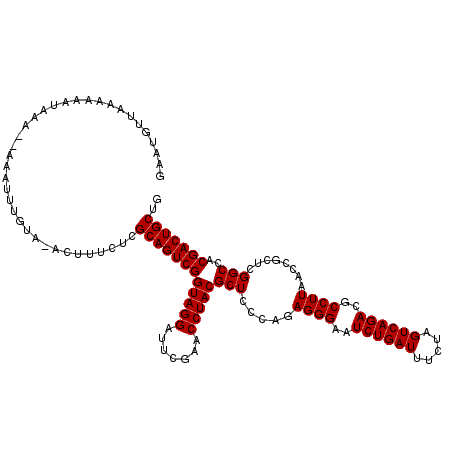
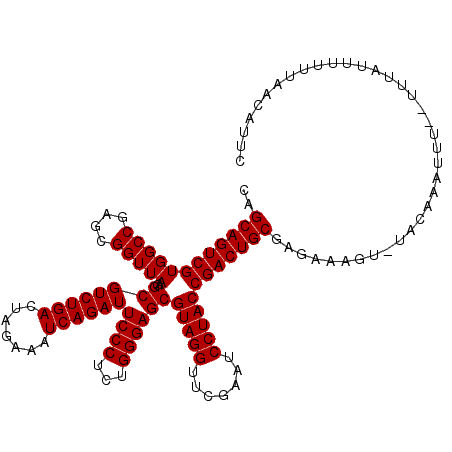
| Location | 3,172,966 – 3,173,086 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.30 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -32.90 |
| Energy contribution | -32.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3172966 120 - 22407834 CAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAGGUUUACGGAUUUUCUUUAUUUUUCAACAUUC ..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))(((((((((....)))))))))............... ( -38.30) >DroSec_CAF1 8530 117 - 1 CAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU-UACAAAUUU--UUUAUUUUUUAACAUUC ..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))..(((.((-((.(((...--.....))).)))).))) ( -35.30) >DroSim_CAF1 8973 117 - 1 CAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU-UACAAAUUU--UUUAUUUUUUAACAUUC ..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))..(((.((-((.(((...--.....))).)))).))) ( -35.30) >DroYak_CAF1 8710 109 - 1 CAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAGAAG-UAAAAUUCU--UU-----UUU---ACUU ..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).....(((-(((((....--.)-----)))---)))) ( -37.50) >consensus CAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAGAAAGU_UACAAAUUU__UUUAUUUUUUAACAUUC ..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))..................................... (-32.90 = -32.90 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:06 2006