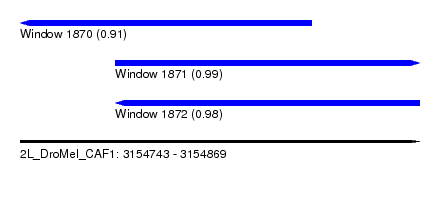
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,154,743 – 3,154,869 |
| Length | 126 |
| Max. P | 0.991171 |

| Location | 3,154,743 – 3,154,835 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
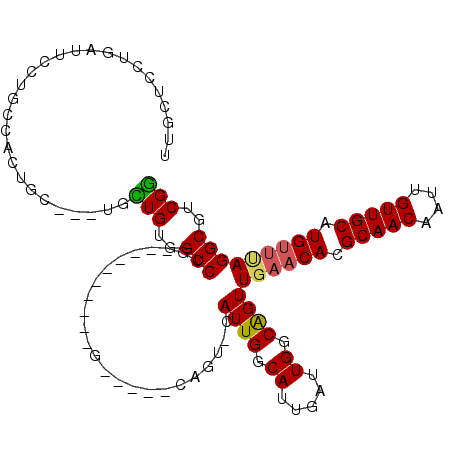
>2L_DroMel_CAF1 3154743 92 - 22407834 GGCAGUCAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGGCAGUGACGGUGA-GACGACAACAACUGCA----CA .(((((..(((.(((((.(((((..(((((((.(((((....))))).)))))))..)))))))))).((....-..)).)))..))))).----.. ( -32.80) >DroVir_CAF1 13114 92 - 1 GUCAGUCAUUGGCAUUGAUUGGCAGCUGAGCACGCAUCAAUUGUUGCAUG-----GCGCUGGCGCUGACGCUGGCGACGCCUCUUGCUGCAGCAGCA ((((((((.......)))))))).((((((((.(((........)))..(-----((((..(((....)))..))...)))...)))).)))).... ( -37.40) >DroEre_CAF1 9807 84 - 1 GGCAGUCAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGGCAGUGACGGUGA-GACGACAAC------------A .....((((((.(((((.(((((..(((((((.(((((....))))).)))))))..)))))))))).))))))-.........------------. ( -28.80) >DroYak_CAF1 12882 87 - 1 GGCAGUCAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGGCAUUGACGGUGA-GACGACAACAA---------CA (.((((((.......)))))).).((((((((.(((((....))))).)))).....((((.(((.....))).-..))))..)))---------). ( -27.60) >DroMoj_CAF1 13538 86 - 1 GUCAGUCAUUGGCAUUGAUUGGCAGCUGAACACGCAACAAUUGUUGCAUG-----GCGCUGGUG------AUGGCGACGCCUCAUGCUGCAGCAGCA ((((((((.......)))))))).((((..((.(((((....))))).))-----((((..(((------(.(((...))))))))).))..)))). ( -32.30) >DroAna_CAF1 10859 92 - 1 AGCAGCCAUUGGCAUUGAUUGGCGGUUGAUCACGCAACAAUUGUUGCAUGUUCAGGCGUCGGCAGUGGCGGUGG-GACGACAUGAGCAGCG----CA .((..((((((.(((((.((((((.((((.((.(((((....))))).)).)))).))))))))))).))))))-(.(.......))...)----). ( -36.70) >consensus GGCAGUCAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGGCAGUGACGGUGA_GACGACAACAGCUGCA____CA .....((((((.((.((.(((((.(((...((.(((((....))))).))....)))))))))).)).))))))....................... (-16.92 = -16.73 + -0.19)


| Location | 3,154,773 – 3,154,869 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -13.78 |
| Energy contribution | -16.06 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3154773 96 + 22407834 CCGACGCCUAAACAUGCAACAAUUGUUGCGUGUUCAACUGCCAAUCAAUGCCAAUG-ACUG-----C----------CGGACCACAGCA---GCAGUGGCAGGAAUCAGGAGCAA ((((..(((.((((((((((....))))))))))...............((((.((-.(((-----(----------.(.....).)))---))).)))))))..)).))..... ( -33.40) >DroEre_CAF1 9829 96 + 1 CCGACGCCUAAACAUGCAACAAUUGUUGCGUGUUCAACUGCCAAUCAAUGCCAAUG-ACUG-----C----------CGGACCACAGCA---GCAGUGGCAGGAAUCAGGAGCAA ((((..(((.((((((((((....))))))))))...............((((.((-.(((-----(----------.(.....).)))---))).)))))))..)).))..... ( -33.40) >DroWil_CAF1 18412 110 + 1 UCGACGCCUGUACAUGCAACAAUUGUUGCAUGUUGAACUGCCAAUCAAUGCCAAUGCACUGGGAUGCAGUGCAAUGAAAGACCACAACA---AAGA--UCAACGAUGACGAUCGA ((((((.......(((((((....)))))))(((((.((((........))((.(((((((.....))))))).)).............---.)).--))))).....)).)))) ( -29.50) >DroYak_CAF1 12907 99 + 1 CCGACGCCUAAACAUGCAACAAUUGUUGCGUGUUCAACUGCCAAUCAAUGCCAAUG-ACUG-----C----------CGGACCACAGCAGCAGCAGUGGCAGGAAUCAGGAGCAA ..(...(((.((((((((((....))))))))))...((((((.....(((...((-.(((-----(----------.(.....).))))))))).)))))).....)))..).. ( -35.00) >DroAna_CAF1 10889 93 + 1 CCGACGCCUGAACAUGCAACAAUUGUUGCGUGAUCAACCGCCAAUCAAUGCCAAUG-GCUG-----C----------UGGACCACAAAA---GCAG---CAGCAAACAGGGCCAA ..(.((..(((.((((((((....)))))))).)))..)).).......(((....-((((-----(----------((..........---.)))---))))......)))... ( -31.60) >consensus CCGACGCCUAAACAUGCAACAAUUGUUGCGUGUUCAACUGCCAAUCAAUGCCAAUG_ACUG_____C__________CGGACCACAGCA___GCAGUGGCAGGAAUCAGGAGCAA ......(((.((((((((((....))))))))))...((((((.....(((.........................................))).)))))).....)))..... (-13.78 = -16.06 + 2.28)

| Location | 3,154,773 – 3,154,869 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3154773 96 - 22407834 UUGCUCCUGAUUCCUGCCACUGC---UGCUGUGGUCCG----------G-----CAGU-CAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGG ..(((.(..(((..(((((.(((---(((((.....))----------)-----))).-)).)))))..)))..).)))((((((.(((((....))))).))))))........ ( -36.10) >DroEre_CAF1 9829 96 - 1 UUGCUCCUGAUUCCUGCCACUGC---UGCUGUGGUCCG----------G-----CAGU-CAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGG ..(((.(..(((..(((((.(((---(((((.....))----------)-----))).-)).)))))..)))..).)))((((((.(((((....))))).))))))........ ( -36.10) >DroWil_CAF1 18412 110 - 1 UCGAUCGUCAUCGUUGA--UCUU---UGUUGUGGUCUUUCAUUGCACUGCAUCCCAGUGCAUUGGCAUUGAUUGGCAGUUCAACAUGCAACAAUUGUUGCAUGUACAGGCGUCGA ((((.((((...((..(--((..---((((((((....))))(((((((.....)))))))..))))..)))..))......(((((((((....)))))))))...)))))))) ( -43.00) >DroYak_CAF1 12907 99 - 1 UUGCUCCUGAUUCCUGCCACUGCUGCUGCUGUGGUCCG----------G-----CAGU-CAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGG .((((..(((((((.(((((.((....)).)))))..)----------)-----.)))-))..)))).....((((..(((((((.(((((....))))).)))))))..)))). ( -36.60) >DroAna_CAF1 10889 93 - 1 UUGGCCCUGUUUGCUG---CUGC---UUUUGUGGUCCA----------G-----CAGC-CAUUGGCAUUGAUUGGCGGUUGAUCACGCAACAAUUGUUGCAUGUUCAGGCGUCGG ...(((.((.((((((---(..(---....)..)..))----------)-----))).-))..)))......(((((.((((.((.(((((....))))).)).)))).))))). ( -33.90) >consensus UUGCUCCUGAUUCCUGCCACUGC___UGCUGUGGUCCG__________G_____CAGU_CAUUGGCAUUGAUUGGCAGUUGAACACGCAACAAUUGUUGCAUGUUUAGGCGUCGG ............................(((..(((........................((((.((.....)).))))((((((.(((((....))))).)))))))))..))) (-16.90 = -16.82 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:58 2006