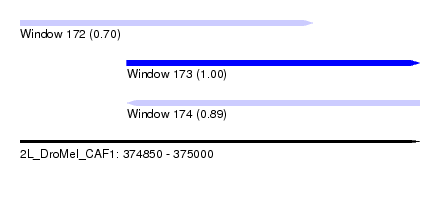
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 374,850 – 375,000 |
| Length | 150 |
| Max. P | 0.999539 |

| Location | 374,850 – 374,960 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 374850 110 + 22407834 CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAAGGCCU ((((((((...(((((....((((.....))))..)))))))))))))..(((..((..(((((..(..(((((((......)--)--------)))))..)..)))))..))..))).. ( -26.60) >DroSec_CAF1 42488 110 + 1 CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAGGCCCU ((((((((...(((((....((((.....))))..)))))))))))))..(((((((...((((..(..(((((((......)--)--------)))))..)..))))))).).)))... ( -26.20) >DroSim_CAF1 43128 110 + 1 CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAGGGCCU ((((((((...(((((....((((.....))))..)))))))))))))..((((.((..(((((..(..(((((((......)--)--------)))))..)..)))))..)).)))).. ( -30.70) >DroEre_CAF1 49067 120 + 1 CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGAACCUGCAUAUACUCGUGUAUACAUAACCAGCCAAGCAGAGGGCCU ((((((((...(((((....((((.....))))..)))))))))))))..((((.((..(((((((((........))))..(((((....)))))........)))))..)).)))).. ( -30.70) >DroYak_CAF1 43621 116 + 1 CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGGCCGCUUAUUGGCCAGGAAUGUGUACGUGCAU--ACUC--GUAUACAUAAUCAGCCAAGCAGAGGGCCU ((((((((...(((((....((((.....))))..)))))))))))))..((((.(((.(((((..(..(((((((((.....--...)--))))))))..)..)))))...))))))). ( -35.20) >consensus CAAUUAGAUUUACGAACAAAUGAAAUUACUUCGCUUUCGUUUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU__A________CACAUAAUCAGCCAAGCAGAGGGCCU ((((((((...(((((....((((.....))))..)))))))))))))..((((.((..(((((..(..(((((....................)))))..)..)))))..)).)))).. (-21.83 = -22.03 + 0.20)

| Location | 374,890 – 375,000 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -34.24 |
| Energy contribution | -34.04 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 374890 110 + 22407834 UUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAAGGCCUUAUCGCUGGCUGGAAAUCUGGCGAUUUUGGGUUGCAUAAA ......(((.(((((...((((.(((((.(((((((......)--)--------)))))..(((((((.((.((........)))))))))))...)))))))))..))))).))).... ( -39.30) >DroSec_CAF1 42528 110 + 1 UUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAGGCCCUUAUCGCUGGCUGGAAAUCUGGCGAUUUUGGGUUGCAUAAA ......(((.(((((...((((.(((((.(((((((......)--)--------)))))..(((((((.((.((........)))))))))))...)))))))))..))))).))).... ( -40.10) >DroSim_CAF1 43168 110 + 1 UUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU--A--------CACAUAAUCAGCCAAGCAGAGGGCCUUAUCGCUGGCUGGAAAUCUGGCGAUUUUGGGUUGCAUAAA ......(((.(((((...((((.(((((.(((((((......)--)--------)))))..(((((((.((.((........)))))))))))...)))))))))..))))).))).... ( -40.10) >DroEre_CAF1 49107 120 + 1 UUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGAACCUGCAUAUACUCGUGUAUACAUAACCAGCCAAGCAGAGGGCCUUAUCGCUGGCUGGAAAUCUGGCGAUUUUGGGUUGCAUAAA ......(((.(((((...((((.(((((.(((((....(((((......))))))))))..(((((((.((.((........)))))))))))...)))))))))..))))).))).... ( -40.90) >DroYak_CAF1 43661 115 + 1 UUUAAUUGCCGGCCGCUUAUUGGCCAGGAAUGUGUACGUGCAU--ACUC--GUAUACAUAAUCAGCCAAGCAGAGGGCCUUAUCGCUGGCUGGAAAUCUGGCGAUU-UGGGUUGCAUAAA ...((((((((((((.....))))).((((((((((((.....--...)--))))))))..(((((((.((.((........)))))))))))...))))))))))-............. ( -40.70) >consensus UUUAAUUGCCGCCCGCUUAUUGGCCAGGAAUGUGUACGUACAU__A________CACAUAAUCAGCCAAGCAGAGGGCCUUAUCGCUGGCUGGAAAUCUGGCGAUUUUGGGUUGCAUAAA ......(((.(((((...((((.(((((.(((((....)))))..................(((((((.((.((........)))))))))))...)))))))))..))))).))).... (-34.24 = -34.04 + -0.20)

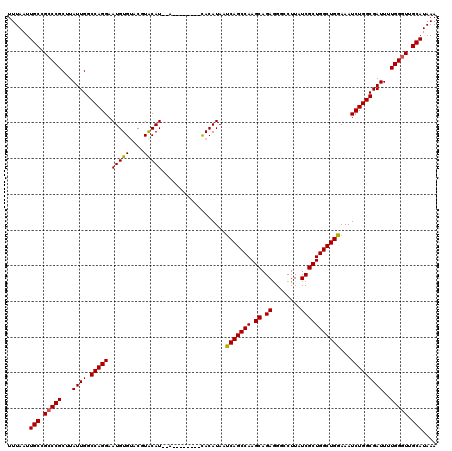
| Location | 374,890 – 375,000 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 374890 110 - 22407834 UUUAUGCAACCCAAAAUCGCCAGAUUUCCAGCCAGCGAUAAGGCCUUCUGCUUGGCUGAUUAUGUG--------U--AUGUACGUACACAUUCCUGGCCAAUAAGCGGGCGGCAAUUAAA ....(((..(((......(((((.....((((((((((........)).)).))))))...(((((--------(--((....))))))))..)))))........)))..)))...... ( -37.64) >DroSec_CAF1 42528 110 - 1 UUUAUGCAACCCAAAAUCGCCAGAUUUCCAGCCAGCGAUAAGGGCCUCUGCUUGGCUGAUUAUGUG--------U--AUGUACGUACACAUUCCUGGCCAAUAAGCGGGCGGCAAUUAAA ....(((..(((......(((((.....((((((((((........)).)).))))))...(((((--------(--((....))))))))..)))))........)))..)))...... ( -36.84) >DroSim_CAF1 43168 110 - 1 UUUAUGCAACCCAAAAUCGCCAGAUUUCCAGCCAGCGAUAAGGCCCUCUGCUUGGCUGAUUAUGUG--------U--AUGUACGUACACAUUCCUGGCCAAUAAGCGGGCGGCAAUUAAA ....(((..(((......(((((.....((((((((((........)).)).))))))...(((((--------(--((....))))))))..)))))........)))..)))...... ( -36.84) >DroEre_CAF1 49107 120 - 1 UUUAUGCAACCCAAAAUCGCCAGAUUUCCAGCCAGCGAUAAGGCCCUCUGCUUGGCUGGUUAUGUAUACACGAGUAUAUGCAGGUUCACAUUCCUGGCCAAUAAGCGGGCGGCAAUUAAA ....(((..(((......(((((((..(((((((((((........)).)).)))))))..))((((((....))))))..............)))))........)))..)))...... ( -35.94) >DroYak_CAF1 43661 115 - 1 UUUAUGCAACCCA-AAUCGCCAGAUUUCCAGCCAGCGAUAAGGCCCUCUGCUUGGCUGAUUAUGUAUAC--GAGU--AUGCACGUACACAUUCCUGGCCAAUAAGCGGCCGGCAAUUAAA ....(((......-.(((((..(........)..)))))..((((..((..(((((((...((((.(((--(.(.--...).)))).))))...)))))))..)).)))).)))...... ( -31.60) >consensus UUUAUGCAACCCAAAAUCGCCAGAUUUCCAGCCAGCGAUAAGGCCCUCUGCUUGGCUGAUUAUGUG________U__AUGUACGUACACAUUCCUGGCCAAUAAGCGGGCGGCAAUUAAA ....(((..(((......(((((.....((((((((((........)).)).))))))...................(((........)))..)))))........)))..)))...... (-26.38 = -26.58 + 0.20)

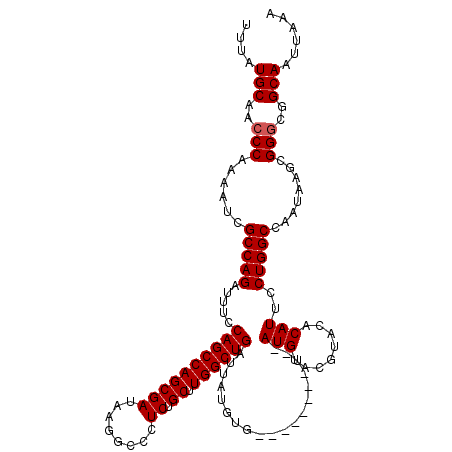
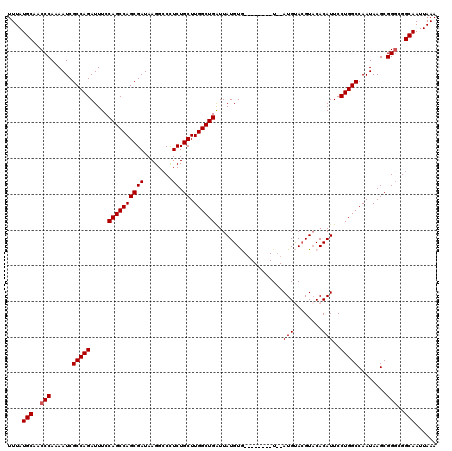
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:17 2006