
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,112,630 – 3,112,735 |
| Length | 105 |
| Max. P | 0.873580 |

| Location | 3,112,630 – 3,112,735 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -10.01 |
| Energy contribution | -11.87 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3112630 105 - 22407834 UUCUUUUCACUUUUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGGCCUGGCCAAGUGCCAUCAAAAAAAAUAAAUAGUCAAAAAAA--AAAAAAACCUGGAAAAU ...(((((.......((((((((((..(((((..((....))..)))))))).)))))))................(((.........--........)))))))). ( -19.53) >DroVir_CAF1 63349 84 - 1 UUC----CGUUUU--AUUGAUGCCACCGGCUACGUUUUAAUUGCUGUCGUCAC---------AAAAAU--AAAAGGGAGA-AAAAA-----AAAAUGGAUGGAAAAA (((----(.((((--(((..((....((((..((.......))..))))...)---------)..)))--)))).)))).-.....-----................ ( -14.00) >DroSec_CAF1 37774 101 - 1 UUCUUUUCACUUUUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGGCCUGGCCAAGUGCCAUCAAAA------AAUAGUCAAAAAAAAAAAAAAAACCUGGAAAAU ...((((((..((((((((((((((..(((((..((....))..)))))))).)))))))........------...................))))..)))))).. ( -19.82) >DroSim_CAF1 38116 96 - 1 UUCUUUUCACUUUUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGGCCUGGCCAAGUGCCAUCAAAA------AAUAGUCAGAAA-----AAAAAACCUGGAAAAU ...(((((.......((((((((((..(((((..((....))..)))))))).)))))))........------......(((...-----.......)))))))). ( -22.50) >DroEre_CAF1 38408 100 - 1 UUCUUUUCACUUCUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGACCUGGCCCAGUGCCAUCAAAU--AAUAAAUAGCAAAAAA-----AAAAAACCUGGAAAAU ...((((((..((((......(((...)))..))))....((((((...((((.....))))......--......))))))....-----........)))))).. ( -12.16) >DroYak_CAF1 39017 97 - 1 UUCUUUUCACUUUUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGGCCUGGCCAAGUGCCAUCAAAC--ACUAAAUAGUAAAAAA-----AAAGA---UGGAAAAU ...((((((((((((((((((((((..(((((..((....))..)))))))).)))))))........--(((....)))....))-----)))).---)))))).. ( -26.30) >consensus UUCUUUUCACUUUUUGCAUUUGCCAAAGGCCAAAGAUCAAUUGCUGGCCUGGCCAAGUGCCAUCAAAA__AAUAAAUAGUCAAAAA_____AAAAAACCUGGAAAAU ...............((((((((((..(((((............))))))))).))))))............................................... (-10.01 = -11.87 + 1.86)

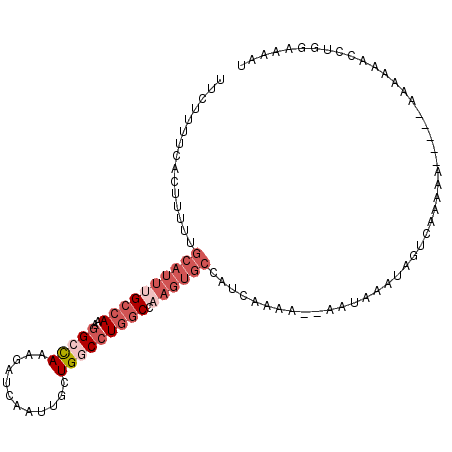
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:44 2006