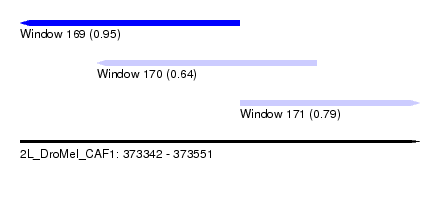
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 373,342 – 373,551 |
| Length | 209 |
| Max. P | 0.952983 |

| Location | 373,342 – 373,457 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 373342 115 - 22407834 UUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAAAAGUUGGCAACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((.....((......))-----.....(((((((.....))))))))))))))((((...(...(((.(((.....))))))..)...))))...................... ( -28.90) >DroSec_CAF1 41018 115 - 1 UUUGCAGCAUUAAAAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAAAAGUUGGCAACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((...........((..-----..)).(((((((.....))))))))))))))((((...(...(((.(((.....))))))..)...))))...................... ( -28.10) >DroSim_CAF1 41678 115 - 1 UUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAAAAGUUGGCAACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((.....((......))-----.....(((((((.....))))))))))))))((((...(...(((.(((.....))))))..)...))))...................... ( -28.90) >DroEre_CAF1 47570 115 - 1 GUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAAAAGUUGGCUACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((.....((......))-----.....(((((((.....))))))))))))))((((...(..((((.(((.....))))))).)...))))...................... ( -31.00) >DroYak_CAF1 42183 120 - 1 GUUGCAGCAUUAAGAUGGGGUUCAACCCAAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAAGGAGCCGAGCGAAAAGUUGGCAACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((.......((((......))))....(((((((.....))))))))))))))((((....(..(((.(((.....))))))..)...))))...................... ( -36.00) >consensus UUUGCAGCAUUAAGAUGGGGUUC_____AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAAAAGUUGGCAACGAGGGGCAUAAAGAUUGAUAACGAGCUAA .(((((((...........(((.......))).(((((((.....))))))))))))))((((....(..(((.(((.....))))))..)...))))...................... (-27.46 = -27.46 + 0.00)

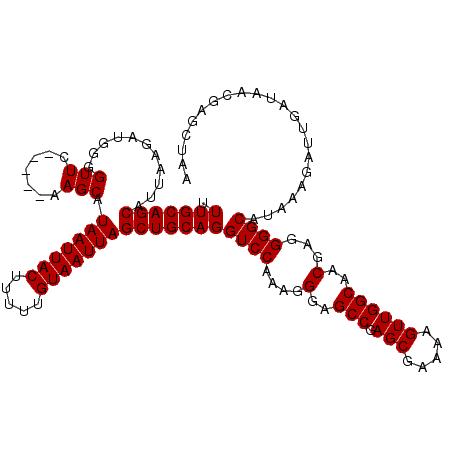
| Location | 373,382 – 373,497 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -36.16 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 373382 115 - 22407834 GGCGAGUACCUCCUGCCCCUUGCGGUUCUUUGUGGCGCCGUUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAA ((((.(.....).)))).....(((((((((.(((.(((...((((((.....((......))-----.....(((((((.....))))))))))))))))))).)))))))))...... ( -38.00) >DroSec_CAF1 41058 115 - 1 GGCGAGUACCUCCUGCCCCUUGCGGUUCUUUGUGGCGCCGUUUGCAGCAUUAAAAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAA ((((.(.....).)))).....(((((((((.(((.(((...((((((...........((..-----..)).(((((((.....))))))))))))))))))).)))))))))...... ( -37.20) >DroSim_CAF1 41718 115 - 1 GGCGAGUACCUCCUGCCCCUUGCGGUUCUUUGUGGCGCCGUUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAA ((((.(.....).)))).....(((((((((.(((.(((...((((((.....((......))-----.....(((((((.....))))))))))))))))))).)))))))))...... ( -38.00) >DroEre_CAF1 47610 115 - 1 GGCGAGUACCGCGUGCCCCUUGCGGUUCUUUGUGGCGCCGGUUGCAGCAUUAAGAUGGGGUUC-----AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAA .(((((.((((((.......))))))..)))))..(((((((((((((.....((......))-----.....(((((((.....)))))))))))))..(((....))))))).))).. ( -38.80) >DroYak_CAF1 42223 120 - 1 GGCAAGUACCUCGUGCCCCUUGCAGUUCUCUGUGGCGCCGGUUGCAGCAUUAAGAUGGGGUUCAACCCAAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAAGGAGCCGAGCGAA ((((.(.....).))))..(..(((....)))..)(((((((((((((.......((((......))))....(((((((.....)))))))))))))..(((....))))))).))).. ( -42.60) >consensus GGCGAGUACCUCCUGCCCCUUGCGGUUCUUUGUGGCGCCGUUUGCAGCAUUAAGAUGGGGUUC_____AAGCAUAAUUACUUUUUGUAAUUAGCUGCAGGUCCAAAGGGAGCCGAGCGAA ((((.(.....).))))..((((((((((((.(((.(((...((((((...........(((.......))).(((((((.....))))))))))))))))))).))))))))..)))). (-36.16 = -36.40 + 0.24)

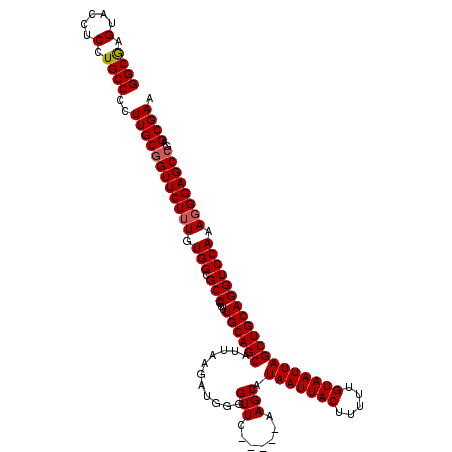
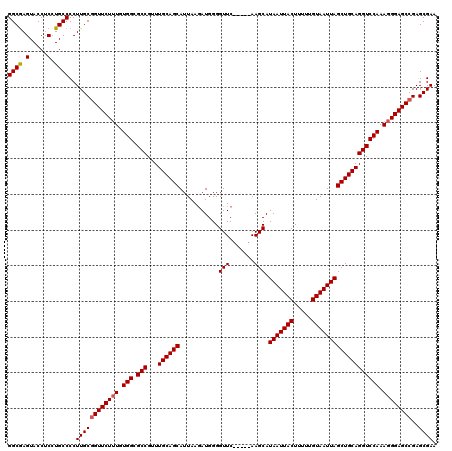
| Location | 373,457 – 373,551 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -22.01 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
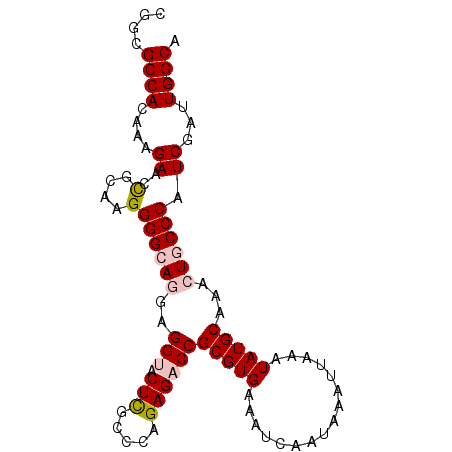
>2L_DroMel_CAF1 373457 94 + 22407834 CGGCGCCACAAAGAACCGCAAGGGGCAGGAGGUACUCGCCCAGAGACCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUGCCCAUCGAUUGGCA ....((((....((..(....)((((((..(((.(((.....))))))(((((................)))))...)))))).))...)))). ( -30.29) >DroSec_CAF1 41133 94 + 1 CGGCGCCACAAAGAACCGCAAGGGGCAGGAGGUACUCGCCGAGAGACCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUGCCCAUCGAUUGGCA ....((((....((..(....)((((((..(((.(((.....))))))(((((................)))))...)))))).))...)))). ( -30.29) >DroSim_CAF1 41793 94 + 1 CGGCGCCACAAAGAACCGCAAGGGGCAGGAGGUACUCGCCCAGAGACCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUGCCCAUCGAUUGGCA ....((((....((..(....)((((((..(((.(((.....))))))(((((................)))))...)))))).))...)))). ( -30.29) >DroEre_CAF1 47685 94 + 1 CGGCGCCACAAAGAACCGCAAGGGGCACGCGGUACUCGCCCAGAGUCCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUACCCAUCGAUUGGCA ....((((....((.((....))..(((((((.((((.....)))))))))))...............................))...)))). ( -30.60) >DroYak_CAF1 42303 94 + 1 CGGCGCCACAGAGAACUGCAAGGGGCACGAGGUACUUGCCCAGAGUCCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUACCCAUCGAUUGGCA ....(((((((....)))...(((.((((.((.((((.....)))))).)))).(((......)))..............)))......)))). ( -20.80) >consensus CGGCGCCACAAAGAACCGCAAGGGGCAGGAGGUACUCGCCCAGAGACCGCGUGAAAUCAAUAAAUUAAAUAUGCAAACUGCCCAUCGAUUGGCA ....((((....((..(....)((((((..(((.(((.....))))))(((((................)))))...)))))).))...)))). (-22.01 = -22.89 + 0.88)

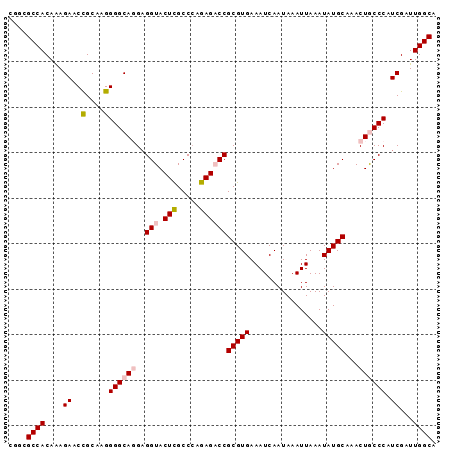
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:14 2006