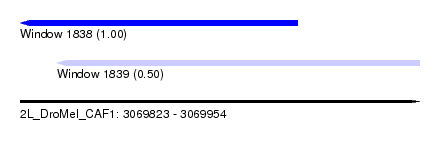
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,069,823 – 3,069,954 |
| Length | 131 |
| Max. P | 0.999782 |

| Location | 3,069,823 – 3,069,914 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.17 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3069823 91 - 22407834 AACUCGGAGGACGAAAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCCGCCUCCAGCACCGCAUCGGGUGGAUCCCUAUCCA .....(((((..(((((((((.((.((((....))))......)).)))))).)))..)))))...........((((((....)))))). ( -33.00) >DroSec_CAF1 125 91 - 1 AACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCUGCCUCCAGCACCGCAUCGGGUGGGUCCCUAUCCA .....(((((..(((((((((.((.((((....))))......)).)))))))))...)))))...........((((((....)))))). ( -38.60) >DroSim_CAF1 66 91 - 1 AACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCCGCCUCCAGCACCGCAUCGGGUGGAUCCCUAUCCA .....(((((..(((((((((.((.((((....))))......)).)))))))))...)))))...........((((((....)))))). ( -37.20) >DroEre_CAF1 68 91 - 1 AACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCCUCCGCCUCCAGCACAGCAUCGGGCGGUUCCCUAUCCA .....(((((..(((((((((.((.((((....))))......)).)))))).)))..)))))...........((..((....))..)). ( -31.90) >DroYak_CAF1 66 91 - 1 AACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCAUCCGCCUCCAGCACAGCAUCGGGCGGAUCCCUAUCCA .....((((((...(((((((.((.((((....))))......)).))))))).(((((((...((...))...))))))))))...))). ( -35.00) >DroAna_CAF1 2289 91 - 1 AAUUCGGAGGACGAAAGCUGCUCAGCCUCCUCGGAGGAUAUGAUGCGCAGCUCCUCUGCUUCCAGCACCGCUUCGGUGGGAUCCCUCACCA .....(((((..((.(((((((((.((((....))))...)))...))))))..))..)))))...........((((((....)))))). ( -36.00) >consensus AACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCCGCCUCCAGCACCGCAUCGGGUGGAUCCCUAUCCA .....(((((..(((((((((.((.((((....))))......)).))))))).))..)))))...........((((((....)))))). (-34.36 = -34.17 + -0.19)

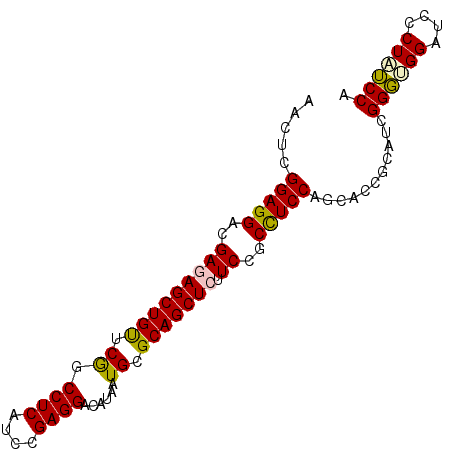
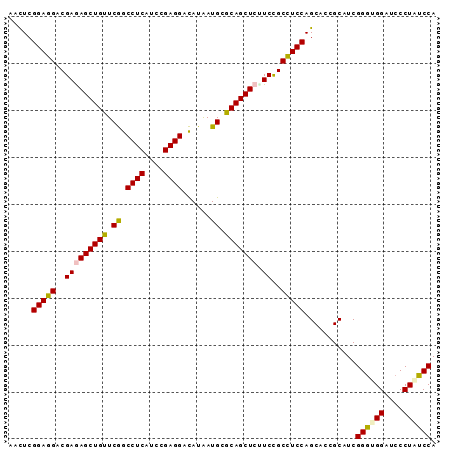
| Location | 3,069,835 – 3,069,954 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -40.62 |
| Energy contribution | -39.27 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3069835 119 - 22407834 AAUCCAGGAGUCCGCCGGCGAGGCCAUGGUCUCCGGUGCUAACUCGGAGGACGAAAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCCGCCUCCAGCACCGCAUCGGGUG- .((((.((((.(((((.....)))...)).))))((((((.....(((((..(((((((((.((.((((....))))......)).)))))).)))..))))))))))).....)))).- ( -52.30) >DroSec_CAF1 137 119 - 1 AAUCCAGGAGUCCGCCGGCGAGGCCAUGGUCUCCGGUGCUAACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCUUCUGCCUCCAGCACCGCAUCGGGUG- .((((.((((.(((((.....)))...)).))))((((((.....(((((..(((((((((.((.((((....))))......)).)))))))))...))))))))))).....)))).- ( -56.50) >DroEre_CAF1 80 119 - 1 AAUCCAGGAGUCCGCCGGCGAGGCCAUGGUCUCCGGUGCCAACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCCUCCGCCUCCAGCACAGCAUCGGGCG- ..(((.((((.(((((.....)))...)).))))(((((...((.(((((..(((((((((.((.((((....))))......)).)))))).)))..)))))))....))))))))..- ( -50.90) >DroWil_CAF1 221 120 - 1 GAUUCAAGAGUCUGCUGGCGAGGCCAUGGUCUCCGGUGCCAAUUCGGAAGAUGAAAGUUGCUCAGCCUCAUCGGAGGAAAUGAUGCGCAGCUCCUCAGCAUCGAGCACGACAUCGGGUAA .......(((((((((...(((((....)))))((((((....((....))(((.(((((((((.((((....))))...)))...))))))..))))))))))))).))).))...... ( -42.70) >DroYak_CAF1 78 119 - 1 AAUUCAGGAGUCCGCCGGCGAGGCUAUGGUCUCCGGUGCUAACUCGGAGGACGAGAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCAUCCGCCUCCAGCACAGCAUCGGGCG- ............((((...(((((....)))))(((((((..((.(((((..(((((((((.((.((((....))))......)).))))))).))..)))))))...)))))))))))- ( -50.70) >DroAna_CAF1 2301 119 - 1 GAUCCAGGAAUCCGCCGGUGAGGCAAUGGUCUCCGGCGCCAAUUCGGAGGACGAAAGCUGCUCAGCCUCCUCGGAGGAUAUGAUGCGCAGCUCCUCUGCUUCCAGCACCGCUUCGGUGG- ......((((..((((((..((((....))))))))))......(((((((.....((((((((.((((....))))...)))...)))))))))))).))))....((((....))))- ( -50.50) >consensus AAUCCAGGAGUCCGCCGGCGAGGCCAUGGUCUCCGGUGCCAACUCGGAGGACGAAAGCUGUUCGGCCUCAUCCGAGGACAUAAUGCGCAGCUCCUCCGCCUCCAGCACCGCAUCGGGUG_ ...((..((((.((((((..((((....))))))))))...))))(((((.....((((((.((.((((....))))......)).))))))......)))))...........)).... (-40.62 = -39.27 + -1.35)
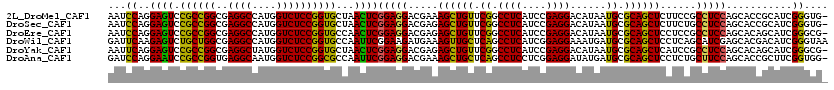
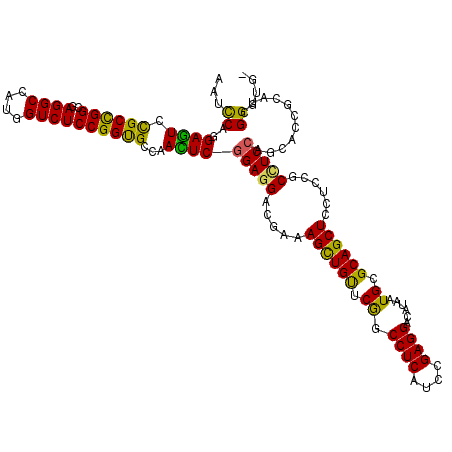
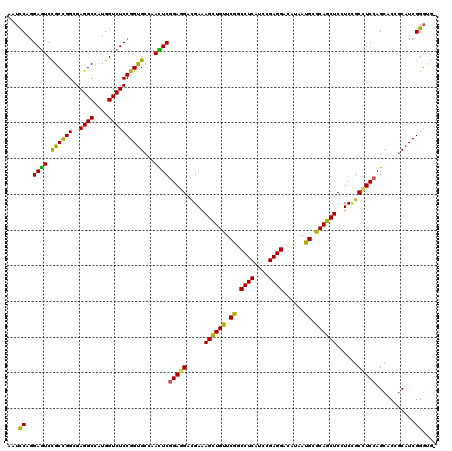
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:26 2006