| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,048,696 – 3,048,898 |
| Length | 202 |
| Max. P | 0.996684 |

| Location | 3,048,696 – 3,048,787 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3048696 91 + 22407834 ----------------------------AGGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAC- ----------------------------...........(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......- ( -19.90) >DroSec_CAF1 1701 91 + 1 ----------------------------AAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAC- ----------------------------...........(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......- ( -19.90) >DroSim_CAF1 2110 90 + 1 -----------------------------AGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAC- -----------------------------..........(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......- ( -19.90) >DroEre_CAF1 2236 119 + 1 GAGUCCUAUCUCCCUUGGCUAUGUGAUCAAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAC- (((......))).((((((.....).)))))........(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......- ( -23.90) >DroYak_CAF1 1739 92 + 1 ----------------------------AAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGACACUCUAAUGAGUCUAAACG ----------------------------..(((......((..(((((((.....((...((((((((.(.......).)))))))).)))))))))..))((((....))))...))). ( -20.60) >DroAna_CAF1 1657 93 + 1 --------------------------GGAAAGUACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAAC- --------------------------.............(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))......- ( -22.30) >consensus ____________________________AAGUUACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAC_ .......................................(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))....... (-20.65 = -20.23 + -0.41)

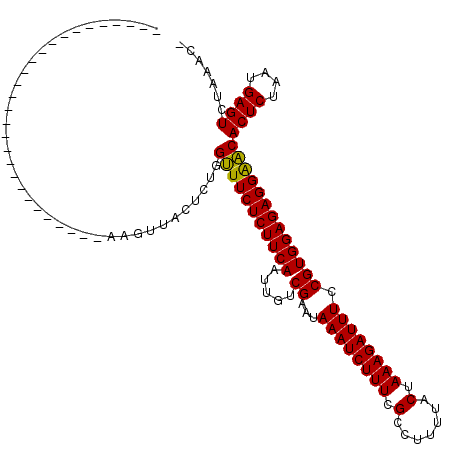
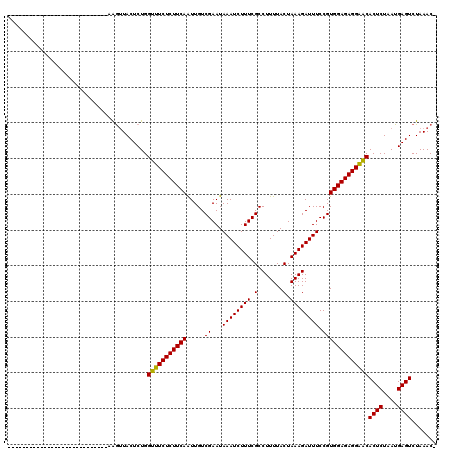
| Location | 3,048,787 – 3,048,898 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.36 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3048787 111 + 22407834 -ACAAUUUUUGCUUAGAGCCCCGAUGGCAUUUGCCUUUGGGGCCAUAAAUGAUGCAUUUCAAAACCAAAAUAUC-CCAUUCGUAUUCUUUUUAAUU-------CUCUUCUUUCAUUAUUA -.....(((((.(((..(((((((.(((....))).)))))))..))).(((......)))....)))))....-.....................-------................. ( -21.70) >DroSec_CAF1 1792 102 + 1 -ACAAUUUUUGCUUAGAGCCCCGAUGGCGUUUGCCUUUGGGGCCAUAAAUGAUACAUU-------CAAAAUAUC-CCAUUUAUAUUCUU--UCAUU-------CUCUUCUUUCAUUAUUA -........((...((((((((((.(((....))).))))))).(((((((.......-------.........-.)))))))..))).--.))..-------................. ( -20.93) >DroSim_CAF1 2200 109 + 1 -ACAAUUUUUGCUUAGAGCCCCGAUGGCGUUUGCCUUUGGGGCCAUAAAUAAUACAUUUUAUACCCGAAAUAUC-CCAUUUAUAUUCUU--UCAUU-------CUCUUCUUUCGUUAUUA -.....(((((....(.(((((((.(((....))).))))))))(((((........)))))...)))))....-..............--.....-------................. ( -19.60) >DroYak_CAF1 2311 91 + 1 CUGAAUUUUUGCUUAGAGCCCGACUGGCGUAUGCCUCUGGGGCCAUAAAUGAUAAAUUUUAAU-UCUAACUAU-AGCUUUCGU-----------------------UUCUUAAGUU---- ..(((.....((((((.((((.(..(((....)))..).))))..((((........))))..-.....))).-)))......-----------------------))).......---- ( -17.60) >DroAna_CAF1 1750 108 + 1 -U-AAUUUUUUGUUCGUGCCCUAGAGGAGUAUUCCUCCGGGGCCAUAAUAAAUAAAUUUAAAAACUUAAACUUC--AAUCCUUAGCCU---GAAUUAUGUGAGCUAUUCUUUGAU----- -.-(((((((((((...((((..((((......))))..))))...)))))).)))))..............((--((....(((((.---.........).))))....)))).----- ( -24.30) >consensus _ACAAUUUUUGCUUAGAGCCCCGAUGGCGUUUGCCUUUGGGGCCAUAAAUGAUACAUUUUAAA_CCAAAAUAUC_CCAUUCAUAUUCUU__UAAUU_______CUCUUCUUUCAUUAUUA ...............(.(((((((.(((....))).))))))))............................................................................ (-16.56 = -16.36 + -0.20)

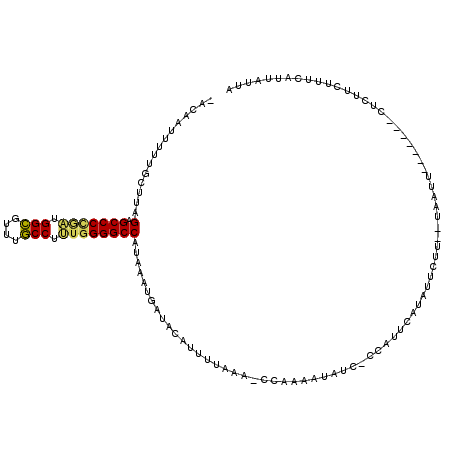
| Location | 3,048,787 – 3,048,898 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
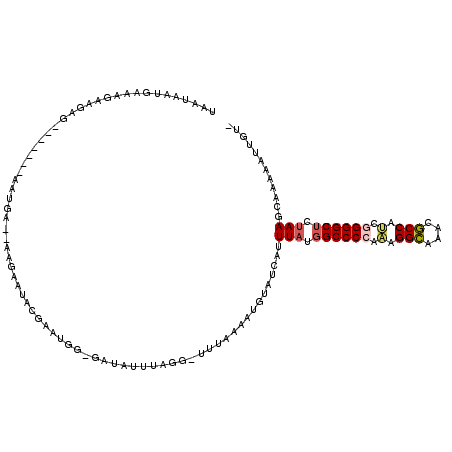
>2L_DroMel_CAF1 3048787 111 - 22407834 UAAUAAUGAAAGAAGAG-------AAUUAAAAAGAAUACGAAUGG-GAUAUUUUGGUUUUGAAAUGCAUCAUUUAUGGCCCCAAAGGCAAAUGCCAUCGGGGCUCUAAGCAAAAAUUGU- ..(((((........((-------(...........((..(((((-..((((((......))))))..)))))..))(((((...(((....)))...))))))))........)))))- ( -24.39) >DroSec_CAF1 1792 102 - 1 UAAUAAUGAAAGAAGAG-------AAUGA--AAGAAUAUAAAUGG-GAUAUUUUG-------AAUGUAUCAUUUAUGGCCCCAAAGGCAAACGCCAUCGGGGCUCUAAGCAAAAAUUGU- ..(((((........((-------(....--.....((((((((.-.((((....-------.))))..))))))))(((((...(((....)))...))))))))........)))))- ( -24.49) >DroSim_CAF1 2200 109 - 1 UAAUAACGAAAGAAGAG-------AAUGA--AAGAAUAUAAAUGG-GAUAUUUCGGGUAUAAAAUGUAUUAUUUAUGGCCCCAAAGGCAAACGCCAUCGGGGCUCUAAGCAAAAAUUGU- ......(....).....-------..((.--.(((.(((((((((-.((((((........)))))).)))))))))(((((...(((....)))...))))))))...))........- ( -24.00) >DroYak_CAF1 2311 91 - 1 ----AACUUAAGAA-----------------------ACGAAAGCU-AUAGUUAGA-AUUAAAAUUUAUCAUUUAUGGCCCCAGAGGCAUACGCCAGUCGGGCUCUAAGCAAAAAUUCAG ----.......(((-----------------------.(....)..-.........-..............((((.(((((....(((....)))....))))).))))......))).. ( -15.50) >DroAna_CAF1 1750 108 - 1 -----AUCAAAGAAUAGCUCACAUAAUUC---AGGCUAAGGAUU--GAAGUUUAAGUUUUUAAAUUUAUUUAUUAUGGCCCCGGAGGAAUACUCCUCUAGGGCACGAACAAAAAAUU-A- -----...........(.((.((((((..---....((((((((--........)))))))).........))))))((((..(((((....)))))..))))..)).)........-.- ( -24.46) >consensus UAAUAAUGAAAGAAGAG_______AAUGA__AAGAAUACGAAUGG_GAUAUUUAGG_UUUAAAAUGUAUCAUUUAUGGCCCCAAAGGCAAACGCCAUCGGGGCUCUAAGCAAAAAUUGU_ ........................................................................(((.((((((.(.(((....))).).)))))).)))............ (-14.00 = -14.44 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:22 2006