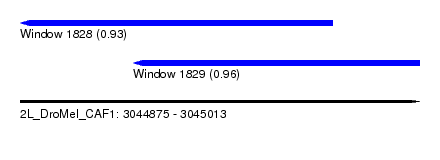
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,044,875 – 3,045,013 |
| Length | 138 |
| Max. P | 0.958404 |

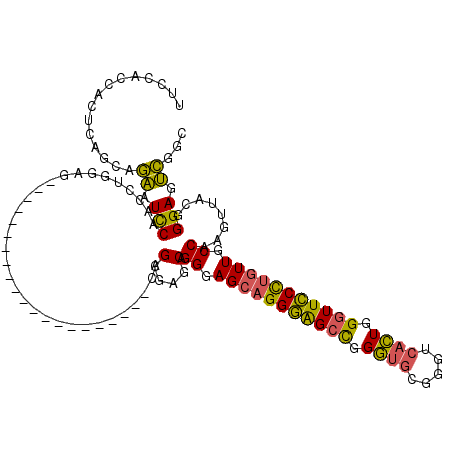
| Location | 3,044,875 – 3,044,983 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -49.38 |
| Consensus MFE | -38.00 |
| Energy contribution | -38.84 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3044875 108 - 22407834 -----------CAGCAGGCAGGGAGCAGGGAGCUGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGCUCUCGAGAUAGUC-GUCCAGCUCUCGACUGCGUGCACUGC -----------..(((.(((((((((((((((((.((((.....)))).))))))))))).))......(((((((.((....(((.....))-).)).))))))).)))).)))..... ( -53.60) >DroSec_CAF1 4613 108 - 1 -----------CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGCUCUCGAGAUAGUC-GUCCAGCUCUCGACUGCGUGCACUGC -----------..(((.(((((((((((((((((.((((.....)))).))))))))))).))......(((((((.((....(((.....))-).)).))))))).)))).)))..... ( -52.60) >DroSim_CAF1 4741 108 - 1 -----------CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGCUCUCGAGAUAGUC-GUCCAGCUCUCGACUGCGUGCACUGC -----------..(((.(((((((((((((((((.((((.....)))).))))))))))).))......(((((((.((....(((.....))-).)).))))))).)))).)))..... ( -52.60) >DroYak_CAF1 4685 119 - 1 AAAGCAGGAAACAGGGAGCUGGGAGCAGGGAGCUGGGUGUGGGUCACUGGGUUCCCUGUUGCCAAGUCACGGGAGUCGGCUCUCGAGAUAGUC-GUCCAGCUCUCGACUGCGUGCACUGC ...(((.(.....(((((((((((((((((((((.((((.....)))).))))))))))).....(((.((((((....)))))).)))....-.)))))))))).....).)))..... ( -60.30) >DroAna_CAF1 4574 99 - 1 -------------------AGGGAG--GGAGGCCCGAUACGGAUCUUUUGAUUUCUUGUUGCCAGGUUGUGGGAGAAGGCUAUCGAGAUAGCCCAGACACCUCGCAACUGCGUGAACUGC -------------------.(((..--.....)))..((((((((....))))....(((((.((((.((..(....((((((....)))))))..)))))).)))))..))))...... ( -27.80) >consensus ___________CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGCUCUCGAGAUAGUC_GUCCAGCUCUCGACUGCGUGCACUGC .............(((.((.((.(((((((((((.((((.....)))).))))))))))).)).((((.((((((....)))))).((.(((.......))).)))))))).)))..... (-38.00 = -38.84 + 0.84)

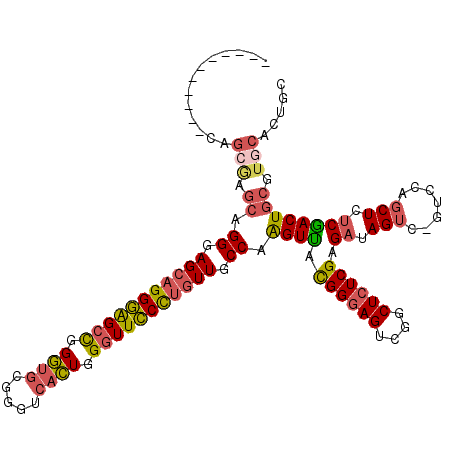
| Location | 3,044,914 – 3,045,013 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -28.75 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3044914 99 - 22407834 UUCCACCACUCAGCAGAAUCCAACCUGAAG---------------------CAGCAGGCAGGGAGCAGGGAGCUGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGC ............((.((.(((..((((...---------------------...))))..((.(((((((((((.((((.....)))).))))))))))).))........))).)).)) ( -40.40) >DroSec_CAF1 4652 99 - 1 UUCCACCACUCAGUAGAAUCCAACCUGGAG---------------------CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGC .....((((((.((((..(((.....)))(---------------------(.....)).((.(((((((((((.((((.....)))).))))))))))).))...))))..)))).)). ( -43.30) >DroSim_CAF1 4780 100 - 1 UUCCAACACUCAGUAGAAUCCAACCUGGAGC--------------------CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGC ((((....(((.((.(..(((.....)))..--------------------).)))))..((.(((((((((((.((((.....)))).))))))))))).)).......))))...... ( -41.70) >DroYak_CAF1 4724 120 - 1 UUCGGCCACUCAGCAGAAUCCAAAACGGAGCAGAAAACAGAAAGCAGGAAACAGGGAGCUGGGAGCAGGGAGCUGGGUGUGGGUCACUGGGUUCCCUGUUGCCAAGUCACGGGAGUCGGC ...(((..((((((....(((.....))).................(....).....))))))(((((((((((.((((.....)))).))))))))))))))..(((((....)).))) ( -46.50) >DroAna_CAF1 4614 87 - 1 UUCGCCAAUUCGACAUUUUUCGAAUUAU-------------------------------AGGGAG--GGAGGCCCGAUACGGAUCUUUUGAUUUCUUGUUGCCAGGUUGUGGGAGAAGGC ...((((((((((......)))))))((-------------------------------((((((--.(((..(((...)))..)))....))))))))..(((.....))).....))) ( -20.50) >consensus UUCCACCACUCAGCAGAAUCCAACCUGGAG_____________________CAGCGAGCAGGGAGCAGGGAGCCGGGUGCGGGUCACUGGGUUCCCUGUUGCCAAGUUACGGGAGUCGGC ...............((.(((................................(....).((.(((((((((((.((((.....)))).))))))))))).))........))).))... (-28.75 = -27.80 + -0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:17 2006