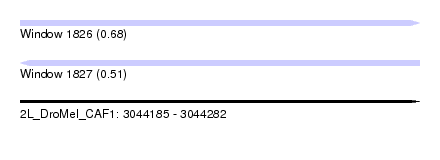
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,044,185 – 3,044,282 |
| Length | 97 |
| Max. P | 0.680274 |

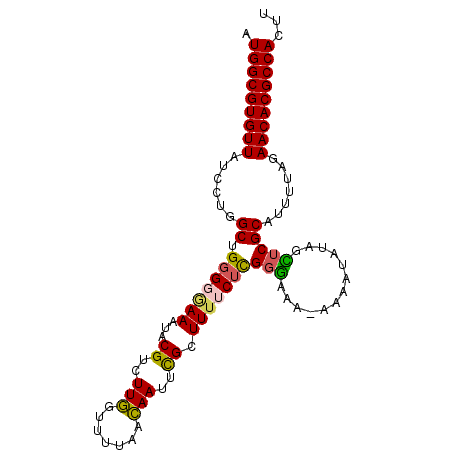
| Location | 3,044,185 – 3,044,282 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3044185 97 + 22407834 AAAUGGCGUGUUCUAAAAUGCGAACUAGAUUUA-UUUUCCGAGAAAAGCGAAUUGUUAAAACCAAGACGUAUUUGACACAGCCAGGAUAACACGCCAU ..((((((((((.......((......(((((.-(((((...)))))..)))))((((((((......)).))))))...))......)))))))))) ( -21.42) >DroSec_CAF1 3913 98 + 1 AAGUGGCGUGUUCUAAAAUGCGAGCUAGAUUUUUUUUCCCGAGAAAAGCAAAUUGUUAAAACCAAGACGUAAUUCCCCCAGCCAGGAUAACACGCCAU ..((((((((((.....((.(..(((.(((((((((((....)))))).)))))(((........)))...........)))..).)))))))))))) ( -21.50) >DroSim_CAF1 3920 97 + 1 AAGUGGCGUGUUCUAAAAUGCGAGCUAUAUUUU-UUUCCCGAGAAAAGCGAAUUGUUAAAACCAAGACGUAAUUCCCCCAGCCAGGAUAACACGCCAU ..((((((((((.....((.(..(((......(-((((....)))))(((..(((.......)))..))).........)))..).)))))))))))) ( -21.70) >DroYak_CAF1 4013 96 + 1 AAGUGGCGUGUUCUGAAAUGCGAACUAU--UUUUUUUCACAAGAAAAGCGAAUUAUUAAAACUAAGACGUAUUUUACCCAGCCAGGAUAACACGCCAU ..((((((((((.(((((((((......--..((((((....)))))).......(((....)))..))))))))).((.....))..)))))))))) ( -23.30) >consensus AAGUGGCGUGUUCUAAAAUGCGAACUAGAUUUU_UUUCCCGAGAAAAGCGAAUUGUUAAAACCAAGACGUAAUUCACCCAGCCAGGAUAACACGCCAU ..((((((((((.......((.............((((....)))).(((..(((.......)))..)))..........))......)))))))))) (-17.59 = -17.10 + -0.50)

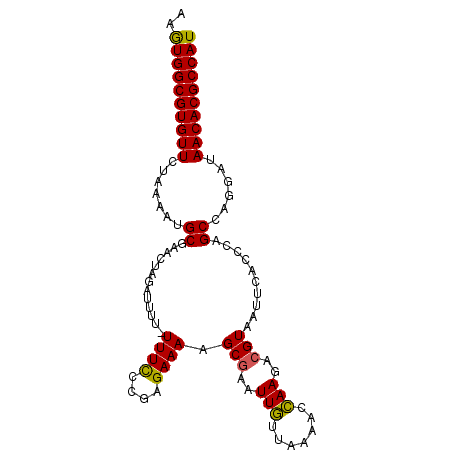
| Location | 3,044,185 – 3,044,282 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3044185 97 - 22407834 AUGGCGUGUUAUCCUGGCUGUGUCAAAUACGUCUUGGUUUUAACAAUUCGCUUUUCUCGGAAAA-UAAAUCUAGUUCGCAUUUUAGAACACGCCAUUU (((((((((..(((((((...))))....((..(((.......)))..))........)))...-....(((((........)))))))))))))).. ( -22.20) >DroSec_CAF1 3913 98 - 1 AUGGCGUGUUAUCCUGGCUGGGGGAAUUACGUCUUGGUUUUAACAAUUUGCUUUUCUCGGGAAAAAAAAUCUAGCUCGCAUUUUAGAACACGCCACUU .((((((((..((((((..(((.(((((................))))).)))...)))))).......(((((........)))))))))))))... ( -24.29) >DroSim_CAF1 3920 97 - 1 AUGGCGUGUUAUCCUGGCUGGGGGAAUUACGUCUUGGUUUUAACAAUUCGCUUUUCUCGGGAAA-AAAAUAUAGCUCGCAUUUUAGAACACGCCACUU .(((((((((......((..((((((...((..(((.......)))..))..))))))(((...-.........))))).......)))))))))... ( -25.42) >DroYak_CAF1 4013 96 - 1 AUGGCGUGUUAUCCUGGCUGGGUAAAAUACGUCUUAGUUUUAAUAAUUCGCUUUUCUUGUGAAAAAAA--AUAGUUCGCAUUUCAGAACACGCCACUU .(((((((((...(((((((((..........))))))........(((((.......))))).....--.............))))))))))))... ( -23.30) >consensus AUGGCGUGUUAUCCUGGCUGGGGGAAAUACGUCUUGGUUUUAACAAUUCGCUUUUCUCGGGAAA_AAAAUAUAGCUCGCAUUUUAGAACACGCCACUU .(((((((((......((.(((((((...((..(((.......)))..)).)))))))(((.............))))).......)))))))))... (-19.33 = -19.27 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:15 2006