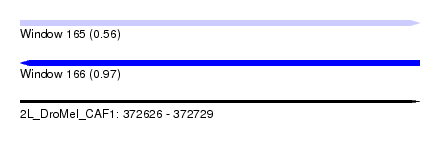
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 372,626 – 372,729 |
| Length | 103 |
| Max. P | 0.969749 |

| Location | 372,626 – 372,729 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -13.92 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 372626 103 + 22407834 UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUUAGCCUUCAUUCCAUCCUCCUUCCGAAAACGGAAAA----------------- ..........((..((.....((((....))))(((((.........))))).................))..))............(((((....)))))..----------------- ( -14.50) >DroSec_CAF1 40287 103 + 1 UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUUGCCCUCCAUUCCAUCCUCCUUCCGAAAACGGAAAA----------------- ..........((..(((....((((....))))(((((.........)))))...............)))..)).............(((((....)))))..----------------- ( -14.00) >DroSim_CAF1 40957 103 + 1 UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUUGCCCUCCAUUCCAUCCUCCUUCCGAAAACGGAAAA----------------- ..........((..(((....((((....))))(((((.........)))))...............)))..)).............(((((....)))))..----------------- ( -14.00) >DroEre_CAF1 46851 103 + 1 UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUAACUCGCCUUUCCAACCUCCUUCCGAAAUCGGAAAA----------------- ..............((.....((((....))))(((((.........)))))....................)).............(((((....)))))..----------------- ( -14.20) >DroYak_CAF1 41483 120 + 1 UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUAACUCGCCACUUCAUCCUCCUUCCGAAAACGAAAAAAGAAGUUGAAUGUUGCA ..............(((.((..(.((....(((((........)))))...............((((...(((....(((...........)))..)))...)))).)).)..)).))). ( -12.90) >consensus UUUAUUAAACGACCGCACCAAGCCACAAAUGGCGAAAAAUAAAUCGCUUUUCACUCUACACAAUCUUUACCCUCCAUUCCAUCCUCCUUCCGAAAACGGAAAA_________________ ..............((.....((((....))))((........))))........................................(((((....)))))................... (-10.36 = -10.56 + 0.20)

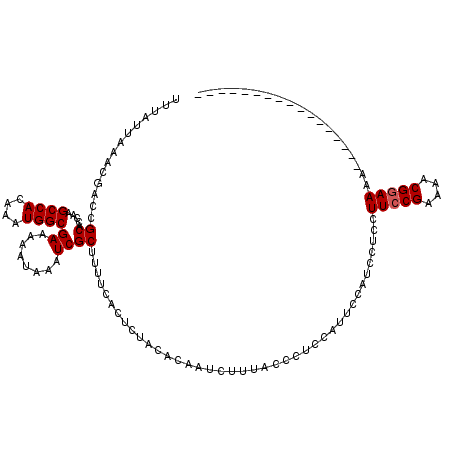
| Location | 372,626 – 372,729 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -26.70 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 372626 103 - 22407834 -----------------UUUUCCGUUUUCGGAAGGAGGAUGGAAUGAAGGCUAAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA -----------------..((((((((((.....))))))))))...........(((((..(.((((.(...((((........))))......).)))).)..))))).......... ( -26.40) >DroSec_CAF1 40287 103 - 1 -----------------UUUUCCGUUUUCGGAAGGAGGAUGGAAUGGAGGGCAAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA -----------------..((((((((((.....))))))))))...........(((((..(.((((.(...((((........))))......).)))).)..))))).......... ( -26.40) >DroSim_CAF1 40957 103 - 1 -----------------UUUUCCGUUUUCGGAAGGAGGAUGGAAUGGAGGGCAAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA -----------------..((((((((((.....))))))))))...........(((((..(.((((.(...((((........))))......).)))).)..))))).......... ( -26.40) >DroEre_CAF1 46851 103 - 1 -----------------UUUUCCGAUUUCGGAAGGAGGUUGGAAAGGCGAGUUAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA -----------------((((((((((((.....)))))))))))).........(((((..(.((((.(...((((........))))......).)))).)..))))).......... ( -27.90) >DroYak_CAF1 41483 120 - 1 UGCAACAUUCAACUUCUUUUUUCGUUUUCGGAAGGAGGAUGAAGUGGCGAGUUAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA .((.((.((((..(((((((((((....))))))))))))))))).)).......(((((..(.((((.(...((((........))))......).)))).)..))))).......... ( -31.30) >consensus _________________UUUUCCGUUUUCGGAAGGAGGAUGGAAUGGAGGGUAAAGAUUGUGUAGAGUGAAAAGCGAUUUAUUUUUCGCCAUUUGUGGCUUGGUGCGGUCGUUUAAUAAA ...................((((((((((.....))))))))))...........(((((..(.((((.(...((((........))))......).)))).)..))))).......... (-26.70 = -25.90 + -0.80)

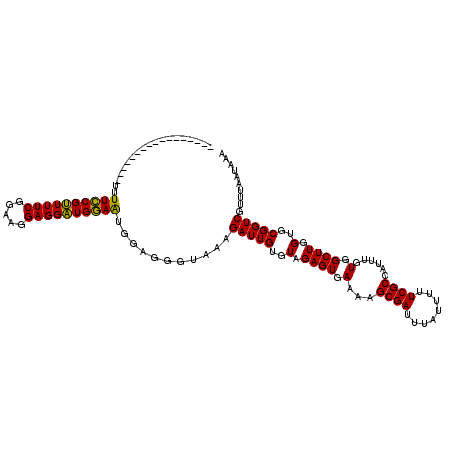
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:09 2006