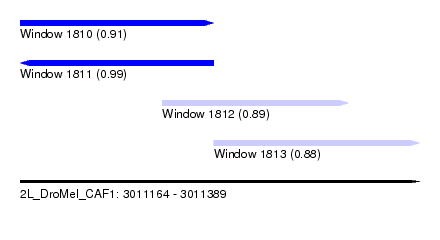
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,011,164 – 3,011,389 |
| Length | 225 |
| Max. P | 0.993562 |

| Location | 3,011,164 – 3,011,273 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -36.77 |
| Energy contribution | -36.66 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3011164 109 + 22407834 AUUGGCAUUUCCGACUGGGAGACCGAGACUCGUCUUCAUUGAAAUCGAGCUCGGAGCUCAAAGCUUAGAGCCUCACAAUCGGACUGGGGACCC-----------GACUGGAUCCCAGUUU .........(((((.((.(((.(((((.((((..(((...)))..))))))))).((((........))))))).)).)))))((((((.((.-----------....)).))))))... ( -42.50) >DroSec_CAF1 18570 109 + 1 AUUGGCAUUUCCGACUGGGAGACCGAGACUCGUCUUCAUUGAAAUCGAGCUCGGAGCUCAAAGCUCAGAGCCCCACAAUCGGACUGGGGACCC-----------GACUGGAUCCCAGUUU .........(((((.((((...(((((.((((..(((...)))..))))))))).((((........))))))))...)))))((((((.((.-----------....)).))))))... ( -42.60) >DroEre_CAF1 19191 120 + 1 AUUGGCAUUUCCGACUGGGAGACCGAGACUCGUCUUCAUUGAAAUCGAGCUCGAAGCUCAGAGCUCAGAGCCUCACAAUCGGACUGGGCACCCAAACGGGGACUGACUGGAUCCCAGUUU ............((((((((..(((((.((((..(((...)))..)))))))).((.((((.((((((..((........)).)))))).(((....)))..)))))))..)))))))). ( -44.80) >consensus AUUGGCAUUUCCGACUGGGAGACCGAGACUCGUCUUCAUUGAAAUCGAGCUCGGAGCUCAAAGCUCAGAGCCUCACAAUCGGACUGGGGACCC___________GACUGGAUCCCAGUUU .........(((((..((....))(((.((((..(((...)))..)))))))((.((((........)))).))....)))))((((((.((................)).))))))... (-36.77 = -36.66 + -0.11)

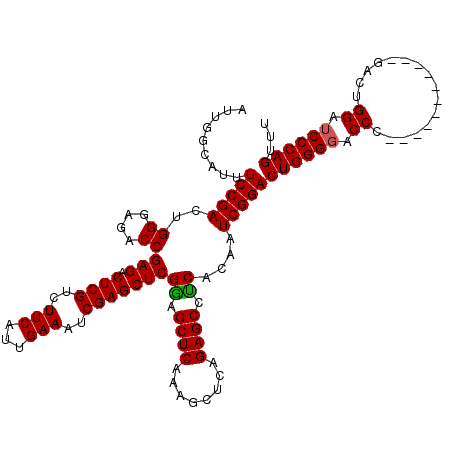
| Location | 3,011,164 – 3,011,273 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -48.33 |
| Consensus MFE | -46.27 |
| Energy contribution | -45.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3011164 109 - 22407834 AAACUGGGAUCCAGUC-----------GGGUCCCCAGUCCGAUUGUGAGGCUCUAAGCUUUGAGCUCCGAGCUCGAUUUCAAUGAAGACGAGUCUCGGUCUCCCAGUCGGAAAUGCCAAU ...(((((((((....-----------)))).)))))((((((((.(((((((........)))).(((((((((.((((...)))).)))).))))).))).))))))))......... ( -48.50) >DroSec_CAF1 18570 109 - 1 AAACUGGGAUCCAGUC-----------GGGUCCCCAGUCCGAUUGUGGGGCUCUGAGCUUUGAGCUCCGAGCUCGAUUUCAAUGAAGACGAGUCUCGGUCUCCCAGUCGGAAAUGCCAAU ...(((((((((....-----------)))).)))))((((((((.(((((...((((.....))))((((((((.((((...)))).)))).))))))))).))))))))......... ( -48.30) >DroEre_CAF1 19191 120 - 1 AAACUGGGAUCCAGUCAGUCCCCGUUUGGGUGCCCAGUCCGAUUGUGAGGCUCUGAGCUCUGAGCUUCGAGCUCGAUUUCAAUGAAGACGAGUCUCGGUCUCCCAGUCGGAAAUGCCAAU ...(((((((((((...........)))))).)))))((((((((.(((((...((((.....))))((((((((.((((...)))).)))).))))))))).))))))))......... ( -48.20) >consensus AAACUGGGAUCCAGUC___________GGGUCCCCAGUCCGAUUGUGAGGCUCUGAGCUUUGAGCUCCGAGCUCGAUUUCAAUGAAGACGAGUCUCGGUCUCCCAGUCGGAAAUGCCAAU ...(((((((((...............)))).)))))((((((((.(((((((........)))).(((((((((.((((...)))).)))).))))).))).))))))))......... (-46.27 = -45.83 + -0.44)

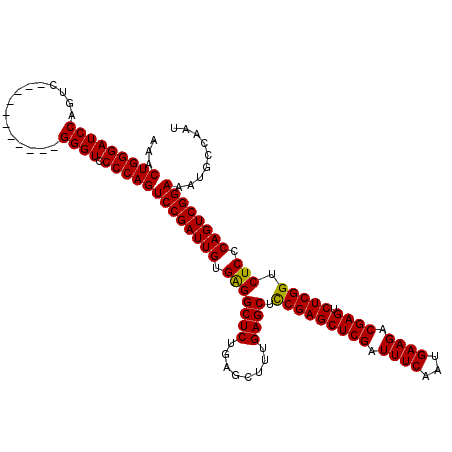
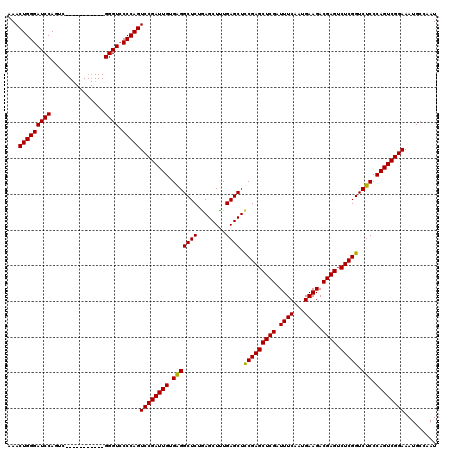
| Location | 3,011,244 – 3,011,349 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -26.39 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3011244 105 + 22407834 GGACUGGGGACCC-----------GACUGGAUCCCAGUUUCAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG----GGCUAACAACAACAA (((((((((.((.-----------....)).)))))))))......((...((((.((((.......))))(((....)))................))))----.))............ ( -29.10) >DroSec_CAF1 18650 105 + 1 GGACUGGGGACCC-----------GACUGGAUCCCAGUUUCAACUUGCACACGGCAUGGCAAUCACAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG----GGCUAACAACAACAA (((((((((.((.-----------....)).)))))))))......((...((((.((((.......))))(((....)))................))))----.))............ ( -29.10) >DroEre_CAF1 19271 120 + 1 GGACUGGGCACCCAAACGGGGACUGACUGGAUCCCAGUUUCAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCGGGCGGGCUAACAACAACAA ((((((((...(((..((....).)..)))..))))))))...(((((...((((.((((.......))))(((....)))................)))).)))))............. ( -33.40) >consensus GGACUGGGGACCC___________GACUGGAUCCCAGUUUCAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG____GGCUAACAACAACAA (((((((((.((................)).)))))))))....((((....((..((((.......))))(((....))).))....))))...(((((.....))))).......... (-26.39 = -26.72 + 0.33)

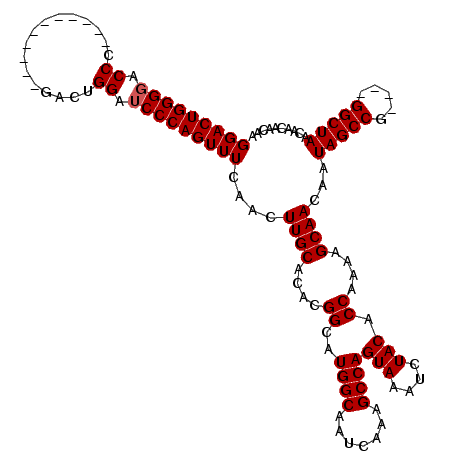
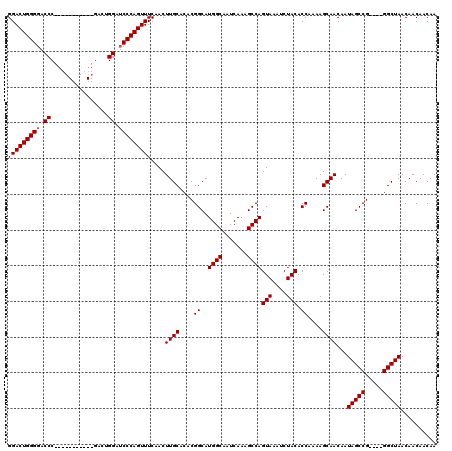
| Location | 3,011,273 – 3,011,389 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -23.93 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3011273 116 + 22407834 CAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG----GGCUAACAACAACAACCAGAGCAAUAUUUGUGCUCAUUAAGCGUAACUGACACAC ......((...((((.((((.......))))(((....)))................))))----..................(((((.......))))).....))............. ( -22.20) >DroSec_CAF1 18679 116 + 1 CAACUUGCACACGGCAUGGCAAUCACAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG----GGCUAACAACAACAACCAGAGCAAUAUUUGUGCUCAUUAAGCGUAACUGACACAC ......((...((((.((((.......))))(((....)))................))))----..................(((((.......))))).....))............. ( -22.20) >DroEre_CAF1 19311 120 + 1 CAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCGGGCGGGCUAACAACAACAACCAGAGCAAUAUUUGUGCUCAUUAAGCGUAACUGACACAC ...(((((...((((.((((.......))))(((....)))................)))).)))))................(((((.......))))).................... ( -25.80) >consensus CAACUUGCACACGGCAUGGCAAUCAAAGCCAGUAAAUCUACACCAAAAGCAACAAUAGCCG____GGCUAACAACAACAACCAGAGCAAUAUUUGUGCUCAUUAAGCGUAACUGACACAC ....((((....((..((((.......))))(((....))).))....((.....(((((.....))))).............(((((.......))))).....))))))......... (-23.93 = -23.93 + 0.00)

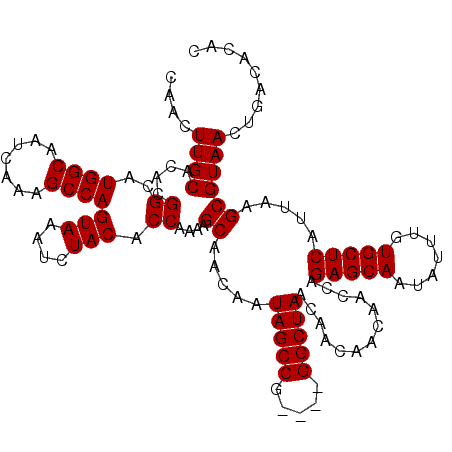
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:01 2006