
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,010,778 – 3,010,898 |
| Length | 120 |
| Max. P | 0.839953 |

| Location | 3,010,778 – 3,010,898 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -24.31 |
| Energy contribution | -24.99 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
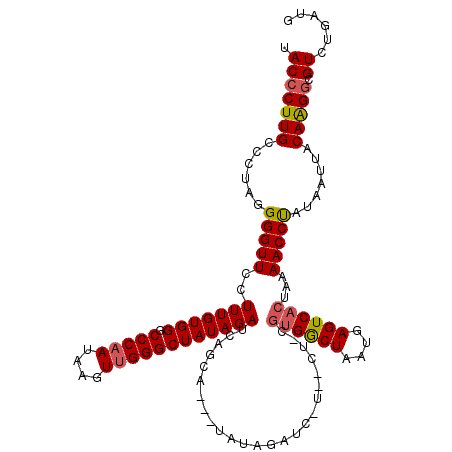
>2L_DroMel_CAF1 3010778 120 + 22407834 UACCCUUGCCCUAGGGGUUCCUUUGUGGGCCCAAUAAGUUGGGCUAUAGAUCAGCAAUCUAUAGAUCUUAGCUGCGUGGCUAAUGAGUCACUUAAACCUAUAAUUACAAGGCGUCUGAUG .(((((((...)))))))..((((((((((((((....))))))(((((..((((.(((....)))....)))).((((((....))))))......))))).))))))))......... ( -35.00) >DroSec_CAF1 18193 99 + 1 UACCCUUGCCCUAGGGGUUGCUUUGUGGGCCCAAUAAGUUGGGCUAUAGAUCAGCA---------------------GACUAAUGAGUCACCAAAACCUAUAAUUACAAGUCGUUUGAUG ..((((......))))((((.(((((((.(((((....)))))))))))).)))).---------------------((((....))))............................... ( -26.90) >DroEre_CAF1 18802 120 + 1 UACCCUUGCCCUAGGGGUUCCUUUGUGGGCCCCAGAAGUUGGGCUAUAGAUCAGCAGAUUAUAGAUCGUGACUUCGUGGCUAAUAAGUCACUGAAACCCAUAAUUACAGGGCGUCUGCUG .......(((((.(((((((......))))))).(((((((((((((.((((....)))))))).)).)))))))((((((....))))))................)))))........ ( -39.20) >consensus UACCCUUGCCCUAGGGGUUCCUUUGUGGGCCCAAUAAGUUGGGCUAUAGAUCAGCA___UAUAGAUC_U__CU_CGUGGCUAAUGAGUCACUAAAACCUAUAAUUACAAGGCGUCUGAUG .(((((((......(((((..(((((((.(((((....)))))))))))).........................((((((....))))))...))))).......))))).))...... (-24.31 = -24.99 + 0.67)

| Location | 3,010,778 – 3,010,898 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
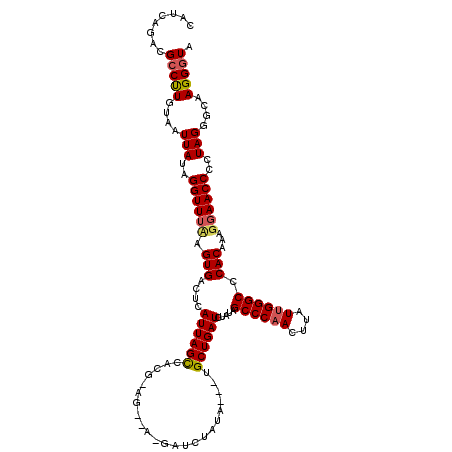
>2L_DroMel_CAF1 3010778 120 - 22407834 CAUCAGACGCCUUGUAAUUAUAGGUUUAAGUGACUCAUUAGCCACGCAGCUAAGAUCUAUAGAUUGCUGAUCUAUAGCCCAACUUAUUGGGCCCACAAAGGAACCCCUAGGGCAAGGGUA ......((.((((((.......(((((..(((..((.(((((......)))))))..((((((((...))))))))((((((....)))))).)))....)))))......)))))))). ( -33.62) >DroSec_CAF1 18193 99 - 1 CAUCAAACGACUUGUAAUUAUAGGUUUUGGUGACUCAUUAGUC---------------------UGCUGAUCUAUAGCCCAACUUAUUGGGCCCACAAAGCAACCCCUAGGGCAAGGGUA ((((((..(((((((....)))))))))))))((((.......---------------------((((........((((((....))))))......)))).(((...)))...)))). ( -26.34) >DroEre_CAF1 18802 120 - 1 CAGCAGACGCCCUGUAAUUAUGGGUUUCAGUGACUUAUUAGCCACGAAGUCACGAUCUAUAAUCUGCUGAUCUAUAGCCCAACUUCUGGGGCCCACAAAGGAACCCCUAGGGCAAGGGUA (((((((.((((.........))))....(((((((..........))))))).........))))))).......((((...((((((((.((.....))...))))))))...)))). ( -40.70) >consensus CAUCAGACGCCUUGUAAUUAUAGGUUUAAGUGACUCAUUAGCCACG_AG__A_GAUCUAUA___UGCUGAUCUAUAGCCCAACUUAUUGGGCCCACAAAGGAACCCCUAGGGCAAGGGUA ........(((((....(((..((((((.(((....((((((.......................)))))).....((((((....)))))).)))...))))))..)))....))))). (-21.35 = -22.13 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:58 2006