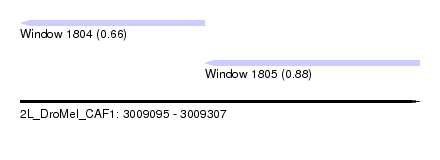
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,009,095 – 3,009,307 |
| Length | 212 |
| Max. P | 0.882535 |

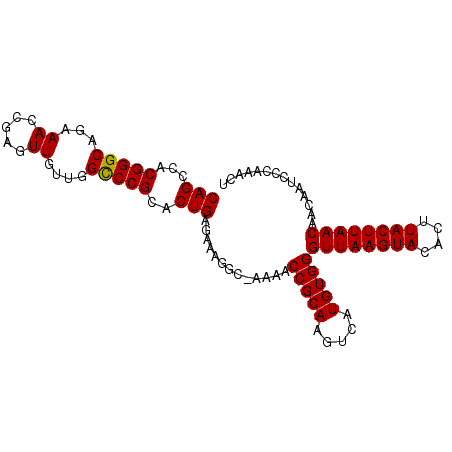
| Location | 3,009,095 – 3,009,193 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -22.19 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3009095 98 - 22407834 CAGCCACGGGCAGAAACCAAGUUGUUGGUCCGCACUGAGACAAGC-AAAACCGCAAGUCAUGUGGGUUAAGUACACUUACUUAACAACAAUCCCAAACU ...((((((((...(((......))).)))).......(((..((-......))..)))..))))((((((((....)))))))).............. ( -24.70) >DroSec_CAF1 16556 98 - 1 CAGCCACGGGCAGAAAGCGAGUUGUUGGCCCGCACUGAGGAAGGC-AAAACCGCAAGUCAUGUGGGUUAAGUAAACUUACUUAACAACAAUCCCAAACU (((...(((((....(((.....))).)))))..))).(((....-....(((((.....)))))((((((((....)))))))).....)))...... ( -28.00) >DroEre_CAF1 17163 99 - 1 CAGCCACGGGCAGAAACCGAGUUUUUAGCCCGCACUGAGAAAGGCCAAAACCGCAAGUCAUGUGGGUUAAGUACACUUACUUAACAACAAUCCCAAACU (((...(((((((((((...)))))).)))))..))).....((......(((((.....)))))((((((((....)))))))).......))..... ( -28.20) >consensus CAGCCACGGGCAGAAACCGAGUUGUUGGCCCGCACUGAGAAAGGC_AAAACCGCAAGUCAUGUGGGUUAAGUACACUUACUUAACAACAAUCCCAAACU (((...(((((...((.....))....)))))..))).............(((((.....)))))((((((((....)))))))).............. (-22.19 = -21.97 + -0.22)

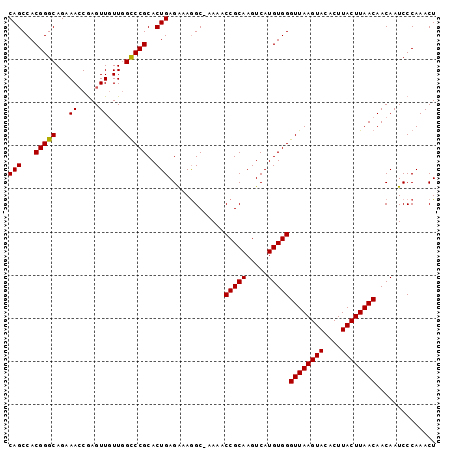
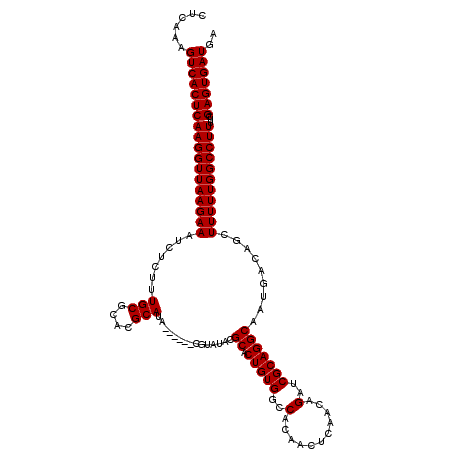
| Location | 3,009,193 – 3,009,307 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3009193 114 - 22407834 CUCAAAGUCACUCAAGGUUAAGAAAUCUCUUUGCGCACGCAUA------CGUAUACGCACUGUGGCCACAACUCAACAGAUCGCAGGCAAUGACAGCUUUUUGGCCUUUUCGAGUGAUGA ......((((((((((((((((((.(.((.(((((((((....------)))....)).(((((..(...........)..))))))))).)).)..)))))))))))...))))))).. ( -32.20) >DroSec_CAF1 16654 108 - 1 CUCAAAGUCACUCAAGGUUAAGAAAUCUCUUUGCGCACGCAU------------ACGCACUGUGGCCACAACUCAACAGAUCGCAGGCAAUGACAGCUUUUUGGCCUUUUCGAGUGAUGA ......((((((((((((((((((.(.((.((((....((..------------..)).(((((..(...........)..))))))))).)).)..)))))))))))...))))))).. ( -32.10) >DroEre_CAF1 17262 120 - 1 CUCAAAGUCACUCAAGGUUAAGAAAUCUCUUUGCGCACGCAUACGCACACGUAUACGCACUGUGGCCACAACUCAACAGAUCGCAGGCAAUGACAGCUUUUUGGCCUUUUCGAGUGAUGA ......((((((((((((((((((.(.((.((((...((.(((((....))))).))..(((((..(...........)..))))))))).)).)..)))))))))))...))))))).. ( -35.50) >consensus CUCAAAGUCACUCAAGGUUAAGAAAUCUCUUUGCGCACGCAUA______CGUAUACGCACUGUGGCCACAACUCAACAGAUCGCAGGCAAUGACAGCUUUUUGGCCUUUUCGAGUGAUGA ......((((((((((((((((((.......(((....)))...............((.(((((..(...........)..))))))).........)))))))))))...))))))).. (-30.90 = -30.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:54 2006