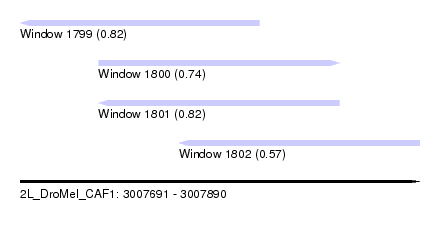
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,007,691 – 3,007,890 |
| Length | 199 |
| Max. P | 0.821660 |

| Location | 3,007,691 – 3,007,810 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -23.37 |
| Energy contribution | -22.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3007691 119 - 22407834 CCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUCCAGCACAGCAAACAGCUGUGGGAAUUCUAAUUUUCAGCUGGCUAAUGGAAAGAAGAAAGGCUUUACGAUAAGGGAUUAAAAA-GUU (((.(((.((((((((...(((((((....)))))))((((((.....))))))))).((((..((((((.........))))))..))))..))))).).)).))).........-... ( -37.80) >DroSec_CAF1 15189 116 - 1 CCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUGCCGCACAGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAUGGAAAGAAGAAAGGCUGUACGAUAAGGGAUUA---A-GUU (((........(((((.....))))).(((....((.((((((.....))))))))............(((((..((..(....)..))...))))))))....))).....---.-... ( -32.60) >DroEre_CAF1 15744 114 - 1 CUUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU------UGGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAUGGAAAGAAGAAAGGGCUUAACGAAAGGGAUUAAAAAGACU .(((((.....(((((.....)))))...((((------(......((((((...((((....)))).)))))).((..(....)..))...)))))...(....)..)))))....... ( -22.80) >consensus CCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU_C_GCACAGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAUGGAAAGAAGAAAGGCUUUACGAUAAGGGAUUAAAAA_GUU (((........(((((.....)))))(((.(((.......)))...((((((...((((....)))).)))))))))...........................)))............. (-23.37 = -22.93 + -0.44)

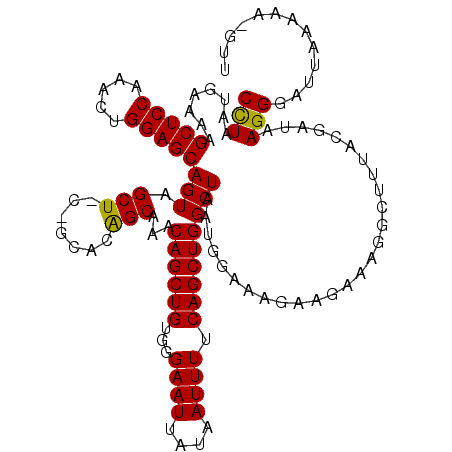
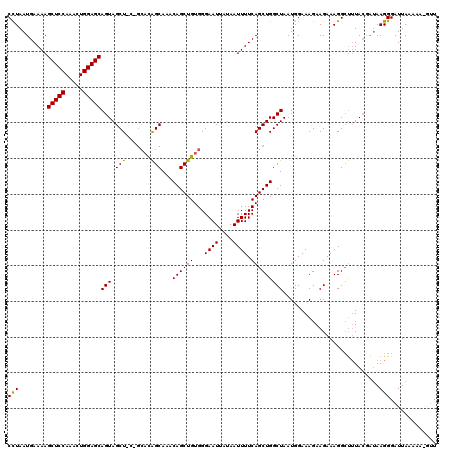
| Location | 3,007,730 – 3,007,850 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.76 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3007730 120 + 22407834 AUUAGCCAGCUGAAAAUUAGAAUUCCCACAGCUGUUUGCUGUGCUGGAGCUACUGCUCCAGUUUGGAGCUUUUCAUUAGGAAAAGCCGCCGCAUUGGUAGUCAACUCAACCCCCACCGAC ....((..((.............(((((((((.....))))))(((((((....)))))))...)))(((((((.....))))))).)).))..(((..((.......))..)))..... ( -36.90) >DroSec_CAF1 15225 120 + 1 AUUAGCCAGCUGAAAAUUAUAAUUCCCACAGCUGUUUGCUGUGCGGCAGCUACUGCUCCAGUUUGGAGCUUUUCAUUAGGAAAAGCCACCGCAUUGGUAAUCAACUUAACCACCAUCAUC ...(((.(((((............((((((((.....)))))).)))))))...)))(((((.(((.(((((((.....)))))))..))).)))))....................... ( -33.00) >DroEre_CAF1 15784 114 + 1 AUUAGCCAGCUGAAAAUUAUAAUUCCCACAGCUGUUUGCCA------AGCUACUGCUCCAGUUUGGAGCUUUUCAUUAAGAAAAGCCGCCGCAUUGGUAAUCAACUUAGCCCCCACCAAC ....((((((((................)))))).((((((------(((.((((...))))..((.(((((((.....)))))))..)))).)))))))........)).......... ( -27.59) >consensus AUUAGCCAGCUGAAAAUUAUAAUUCCCACAGCUGUUUGCUGUGC_G_AGCUACUGCUCCAGUUUGGAGCUUUUCAUUAGGAAAAGCCGCCGCAUUGGUAAUCAACUUAACCCCCACCAAC ..((((((((((................))))))...))))......(((....)))(((((.(((.(((((((.....)))))))..))).)))))....................... (-25.64 = -25.76 + 0.11)

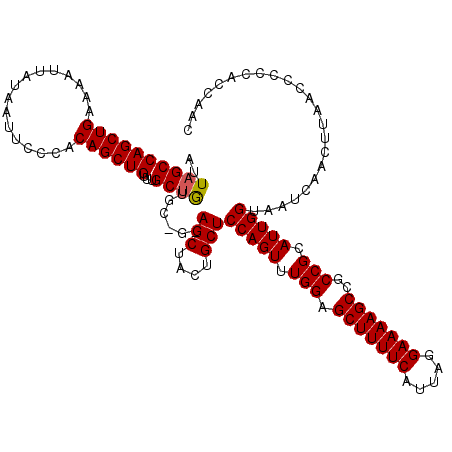
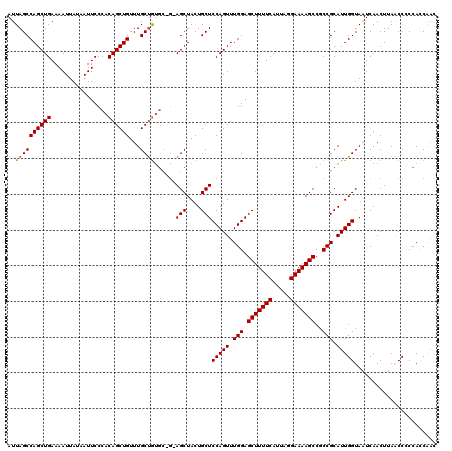
| Location | 3,007,730 – 3,007,850 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -30.00 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3007730 120 - 22407834 GUCGGUGGGGGUUGAGUUGACUACCAAUGCGGCGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUCCAGCACAGCAAACAGCUGUGGGAAUUCUAAUUUUCAGCUGGCUAAU (((((((..(((((((((............((.((((((((.....))))))))))...(((((((....)))))))((((((.....))))))..))))).))))..)).))))).... ( -47.10) >DroSec_CAF1 15225 120 - 1 GAUGAUGGUGGUUAAGUUGAUUACCAAUGCGGUGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUGCCGCACAGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAU .....((((((((.....)))))))).(((((..(((((((.....)))))(((((.....)))))....))..))))).(((...((((((...((((....)))).)))))))))... ( -43.30) >DroEre_CAF1 15784 114 - 1 GUUGGUGGGGGCUAAGUUGAUUACCAAUGCGGCGGCUUUUCUUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU------UGGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAU ((((((....(((((((((....(((.(..((.((((((((.....))))))))))..).))).....)))))------))))...((((((...((((....)))).)))))))))))) ( -37.60) >consensus GUUGGUGGGGGUUAAGUUGAUUACCAAUGCGGCGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU_C_GCACAGCAAACAGCUGUGGGAAUUAUAAUUUUCAGCUGGCUAAU .......(((((((...((.((.(((.(..((.((((((((.....))))))))))..).))))))).))))))).....(((...((((((...((((....)))).)))))))))... (-30.00 = -29.57 + -0.43)


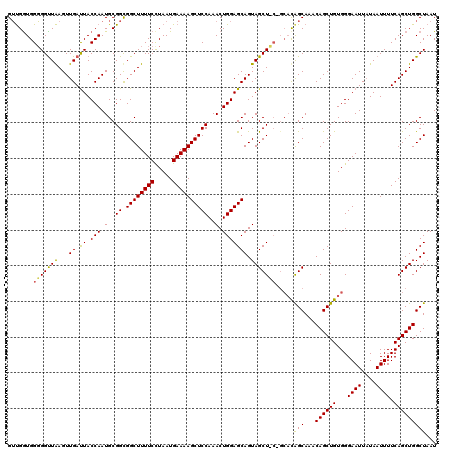
| Location | 3,007,770 – 3,007,890 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -28.86 |
| Energy contribution | -29.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3007770 120 - 22407834 GUUAAAACUGAGUGAAGGUUAAGCGAUAAGUCAAAACUGUGUCGGUGGGGGUUGAGUUGACUACCAAUGCGGCGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUCCAGCAC ...........(((........(((((((((....))).))))(((((..(......)..)))))...))((.((((((((.....))))))))))...(((((((....)))))))))) ( -35.90) >DroSec_CAF1 15265 120 - 1 GUUAAAACUGAGUGAAGGUUAAGCGAUAAGUCAAAACUGUGAUGAUGGUGGUUAAGUUGAUUACCAAUGCGGUGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCUGCCGCAC (((.((.((......)).)).))).....((((......))))..((((((((.....)))))))).(((((..(((((((.....)))))(((((.....)))))....))..))))). ( -35.70) >DroEre_CAF1 15824 114 - 1 GUUAAAACUGAGUGAAGGUUAAGCGAUAAGUCAAAGCUGUGUUGGUGGGGGCUAAGUUGAUUACCAAUGCGGCGGCUUUUCUUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU------U .....((((.......))))((((((.(((((...(((((((((((((...(......).)))))))))))))))))).))..........(((((.....)))))....)))------) ( -35.40) >consensus GUUAAAACUGAGUGAAGGUUAAGCGAUAAGUCAAAACUGUGUUGGUGGGGGUUAAGUUGAUUACCAAUGCGGCGGCUUUUCCUAAUGAAAAGCUCCAAACUGGAGCAGUAGCU_C_GCAC ((((...........(((..((((((....))...(((((((((((((..(......)..))))))))))))).))))..)))........(((((.....)))))..))))........ (-28.86 = -29.53 + 0.67)

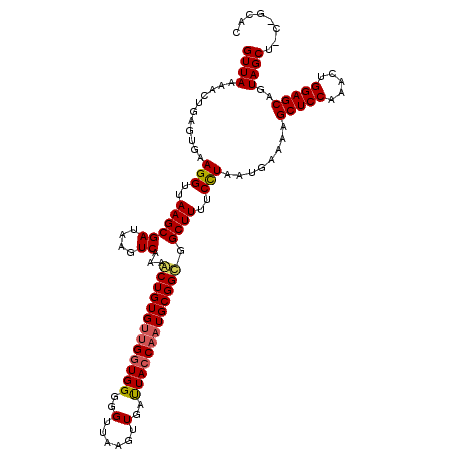
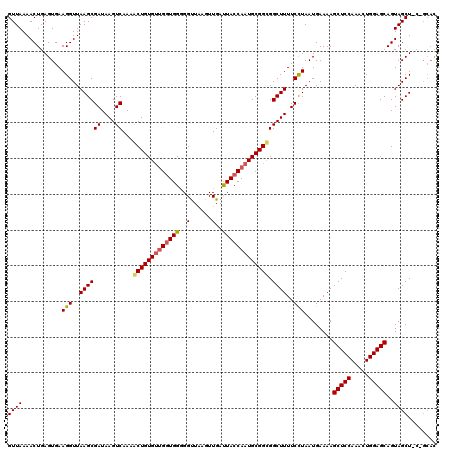
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:52 2006