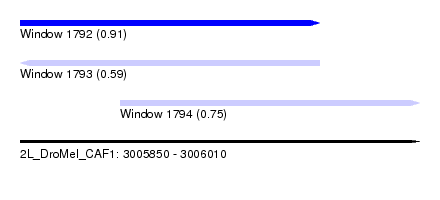
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,005,850 – 3,006,010 |
| Length | 160 |
| Max. P | 0.905070 |

| Location | 3,005,850 – 3,005,970 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005850 120 + 22407834 ACCUUUUAUAGGUAUAAAUCAAUGUGUGACUGUGAUUUAUAUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGAUUGUUCUUUCGGAGAACCCCCUACGGAAAGAAA .(((...((..((((((((((..(.....)..))))))))))..))......))).......((((........))))((((((((...((((((...))))))..)))).))))..... ( -28.40) >DroSec_CAF1 13372 119 + 1 ACCUUUCAUAGGUAUAAAUCAACGUGAGACUGUGAUUUAUAUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGAUCGUUCUCUU-AAGAACCCCCUAGUGAAAGAAG ..(((((((((((((((((((.(....)....)))))))))..(((((...(((((......((((........)))).))))))))))(((((...-.)))))..))).)))))))... ( -34.50) >DroEre_CAF1 13327 116 + 1 ACCUUUCAUAGGUAUAAAUCAACGCGUGACU-UGAUUUAUAUUGAUUCUAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCUUAGGGCCGUUCUCUU-GAGAGC--CCUUGGGGGAGAAA ..((..(...((((((((((((........)-)))))))))))....(((..(((.......((((........))))(((((((((......))))-))))))--)).))))..))... ( -30.90) >DroYak_CAF1 13040 116 + 1 ACCUUUCAUAGGUAUAAAUCAACGUGUGACU-UGAUUUAUAUUGGUUCUAUUGGGAUAAUUUGGUCUAAUUUGUGGCCUUCUUAGGGCUGUUCUCUG-GGGAGC--CCUAGGUAAAGAAA .(((...(((((((((((((((........)-)))))))))).....)))).))).......((((........))))..(((((((((.((.....-)).)))--))))))........ ( -32.40) >consensus ACCUUUCAUAGGUAUAAAUCAACGUGUGACU_UGAUUUAUAUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGACCGUUCUCUU_GAGAAC__CCUAGGGAAAGAAA .(((......(((((((((((...........))))))))))).........))).......((((........))))((((((((...(((((.....)))))..))))))))...... (-25.42 = -25.48 + 0.06)

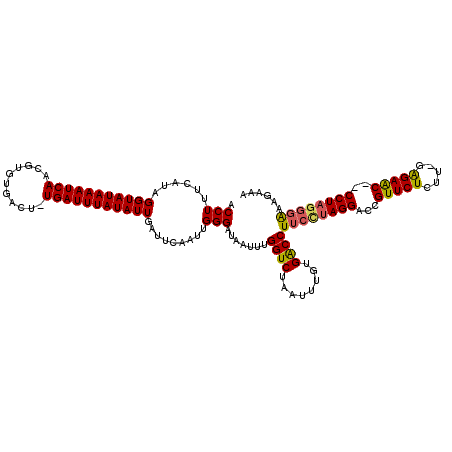
| Location | 3,005,850 – 3,005,970 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005850 120 - 22407834 UUUCUUUCCGUAGGGGGUUCUCCGAAAGAACAAUCCUAGGAAGGUCACAAAUUAGACCAAAUUAUCCCAAUUGAAUCAAUAUAAAUCACAGUCACACAUUGAUUUAUACCUAUAAAAGGU .(((.((((.(((((.(((((.....)))))..)))))))))((((........))))..............)))....(((((((((..(.....)..)))))))))(((.....))). ( -26.00) >DroSec_CAF1 13372 119 - 1 CUUCUUUCACUAGGGGGUUCUU-AAGAGAACGAUCCUAGGAAGGUCACAAAUUAGACCAAAUUAUCCCAAUUGAAUCAAUAUAAAUCACAGUCUCACGUUGAUUUAUACCUAUGAAAGGU ..(((((((.((((((((((((-(.((......)).))))))((((........)))).......)))...........(((((((((..(.....)..)))))))))))))))))))). ( -32.30) >DroEre_CAF1 13327 116 - 1 UUUCUCCCCCAAGG--GCUCUC-AAGAGAACGGCCCUAAGAAGGUCACAAAUUAGACCAAAUUAUCCCAAUAGAAUCAAUAUAAAUCA-AGUCACGCGUUGAUUUAUACCUAUGAAAGGU .((((......(((--(((.((-....))..)))))).....((((........)))).............))))....(((((((((-((.....).))))))))))(((.....))). ( -24.50) >DroYak_CAF1 13040 116 - 1 UUUCUUUACCUAGG--GCUCCC-CAGAGAACAGCCCUAAGAAGGCCACAAAUUAGACCAAAUUAUCCCAAUAGAACCAAUAUAAAUCA-AGUCACACGUUGAUUUAUACCUAUGAAAGGU .((((...(.((((--(((...-........))))))).)..((.(........).)).............))))....(((((((((-(........))))))))))(((.....))). ( -19.30) >consensus UUUCUUUCCCUAGG__GCUCUC_AAGAGAACAACCCUAAGAAGGUCACAAAUUAGACCAAAUUAUCCCAAUAGAAUCAAUAUAAAUCA_AGUCACACGUUGAUUUAUACCUAUGAAAGGU .((((.....((((..(((((.....)))))...))))))))((((........)))).....................(((((((((..(.....)..)))))))))(((.....))). (-15.65 = -16.65 + 1.00)

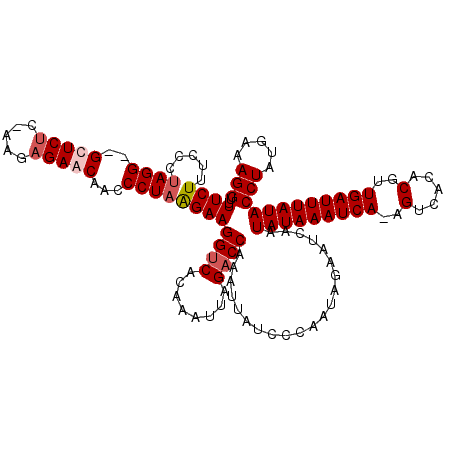
| Location | 3,005,890 – 3,006,010 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.10 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005890 120 + 22407834 AUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGAUUGUUCUUUCGGAGAACCCCCUACGGAAAGAAAUAUGUGCAAGUGCUUGGUUAAUAUUAACAGCUAGCAUCCU ............(((((.....((((........))))((((((((...((((((...))))))..)))).))))..........((.(((..(((((....)))))..))).))))))) ( -27.20) >DroSec_CAF1 13412 106 + 1 AUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGAUCGUUCUCUU-AAGAACCCCCUAGUGAAAGAAGUAUUUGUAAGUUCUUGGUU-------------AGCAUCCU ...(((.((((..(((......((((........))))((((((((...(((((...-.)))))..))))).)))................)))..)))-------------.).))).. ( -24.30) >DroEre_CAF1 13366 117 + 1 AUUGAUUCUAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCUUAGGGCCGUUCUCUU-GAGAGC--CCUUGGGGGAGAAAUAUUUUAAUGUUCGCUGUCAAUAUUAGCAGAUAGCAUCAU ..((((..((((((((((.(((((((........))))(((((((((..(((((...-.)))))--))))))))).))).))))))))))...((((((..........)))))))))). ( -31.70) >DroYak_CAF1 13079 108 + 1 AUUGGUUCUAUUGGGAUAAUUUGGUCUAAUUUGUGGCCUUCUUAGGGCUGUUCUCUG-GGGAGC--CCUAGGUAAAGAAAUAUUUUA---------GUCCAUAUUAGCAGAUAGCAUCAU ..((((.(((((....((((.(((.((((..(((..(.(((((((((((.((.....-)).)))--)))))).)).)..)))..)))---------).))).))))...))))).)))). ( -36.00) >consensus AUUGAUUCAAUUGGGAUAAUUUGGUCUAAUUUGUGACCUUCCUAGGACCGUUCUCUU_GAGAAC__CCUAGGGAAAGAAAUAUUUGAAAGUUCUUGGUCAAUAUUAGCAGAUAGCAUCAU ............(((((.....((((........))))((((((((...(((((.....)))))..)))))))).........................................))))) (-17.21 = -17.77 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:44 2006