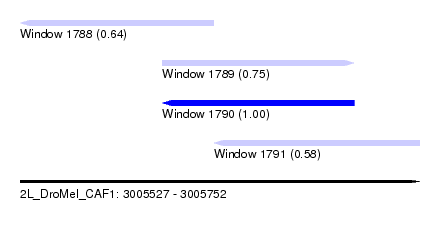
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,005,527 – 3,005,752 |
| Length | 225 |
| Max. P | 0.996064 |

| Location | 3,005,527 – 3,005,636 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -19.01 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005527 109 - 22407834 UGA-----------AUAGGACGUGAGAAGGGCGGUGUGGAAACAAUAAAGAGGCCCGAAAACAAAGCAUUAUGUAAGCAUUAGCACGCUUUAACCUCCCGCACGUCUAGUUAAACAUAAA ..(-----------((.(((((((....(((.(((.((....)).....(((((......(((........)))..((....))..))))).))).))).))))))).)))......... ( -29.00) >DroSec_CAF1 13063 104 - 1 UGA-----------AAAAGACGUGAAAAGGGCGGCGAGGAAACAAUAAAGAGGCCCGAAAACAAAGCAUUAUGUAAGCAUUAGCACGCUUUAAUCUCCCGCACGUCCAGUUAAAC----- ...-----------....((((((....(((.((...(....)......(((((......(((........)))..((....))..)))))..)).))).)))))).........----- ( -21.80) >DroEre_CAF1 12991 106 - 1 CGA-----------AAAAUACGUGAGAAGGGCGGGUAGGAAACAAUAACGAGACCCGAAAACAAAGCGUUAUGUAAGCAUUAGCACGCUUUAACCUCCCGCACGUCCAGUUAAAUAU--- ...-----------.....(((((....((((((((.(..........)...))))).....(((((((.(((....)))....))))))).....))).)))))............--- ( -25.50) >DroYak_CAF1 12694 117 - 1 UGAAAAACUCGUGAAAAAUACGUGAGAAGGGCGGGGAGGUAACAAUAAAGAGAGCCGAAAACAAUGCAUUAUGUAAGCAUUAGCACGCUUUAACCUCCCGCACGUCCAGUUAACUAU--- .......((((((.......))))))..((((((((((((.....(((((.(.((.(....)(((((.........))))).)).).))))))))))))...)))))..........--- ( -28.10) >consensus UGA___________AAAAGACGUGAGAAGGGCGGGGAGGAAACAAUAAAGAGACCCGAAAACAAAGCAUUAUGUAAGCAUUAGCACGCUUUAACCUCCCGCACGUCCAGUUAAACAU___ ..................((((((....(((.((...(....)......................((...(((....)))..)).........)).))).)))))).............. (-19.01 = -19.32 + 0.31)

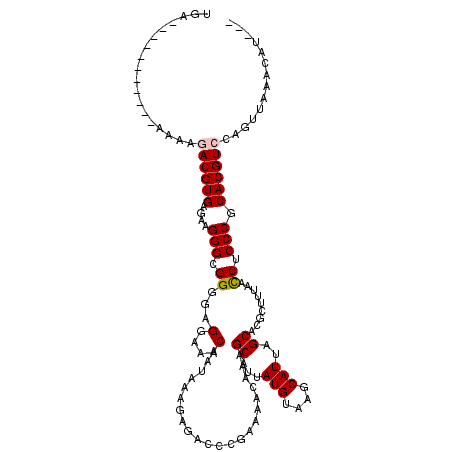
| Location | 3,005,607 – 3,005,715 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -39.01 |
| Consensus MFE | -30.85 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005607 108 + 22407834 UCCACACCGCCCUUCUCACGUCCUAU-----------UCACCUAUU-UUCACGUCUUUUGUACGUGAGGUAGGCCCAGUUGGGCGGAAGGGCGAGAAAGUCGAGGACAUUUUAGGGCCCU ........(((((((((..(((((.(-----------((..(((((-(.((((((....).)))))))))))((((....))))))))))))))))).(((...)))......))))... ( -38.70) >DroSec_CAF1 13138 109 + 1 UCCUCGCCGCCCUUUUCACGUCUUUU-----------UCACGUAUUUUUCACGUCUUCUGUACGUGAGGUAGGCCCAGUUGGGCGGAAGGGCGAGAAAGUCGAGGACAUUUUAGGGUCCU ((((((((((((((((.....((.((-----------((((((((..............)))))))))).))((((....))))))))))))).....).)))))).............. ( -39.44) >DroEre_CAF1 13068 109 + 1 UCCUACCCGCCCUUCUCACGUAUUUU-----------UCGUGUAUUUUCCACGUCUUUUGUACGUGACGUAGGCCCAGUUGGGAGGAAGGGCGAGAAAGUCGAGGACAUUUUAGGGCCUU (((((..((((((((((((((((...-----------.((((.......))))......))))))))......(((....)))..)))))))).....(((...)))....))))).... ( -37.60) >DroYak_CAF1 12771 120 + 1 ACCUCCCCGCCCUUCUCACGUAUUUUUCACGAGUUUUUCAUGUAGUUUUCACGUCUCGUGUACGUGAGGUAGGCCCAGUUGGGUGGAAGGGCGAGAGAGUCGAGGACAUUUUAGGGCCCU ....((((((((((((((((((.....((((((......((((.......))))))))))))))))).....((((....)))).)))))))).(((((((...))).)))).))).... ( -40.30) >consensus UCCUCCCCGCCCUUCUCACGUAUUUU___________UCACGUAUUUUUCACGUCUUUUGUACGUGAGGUAGGCCCAGUUGGGCGGAAGGGCGAGAAAGUCGAGGACAUUUUAGGGCCCU .((....(((((((((...............................((((((((....).)))))))....((((....)))))))))))))((((.(((...))).)))).))..... (-30.85 = -31.10 + 0.25)

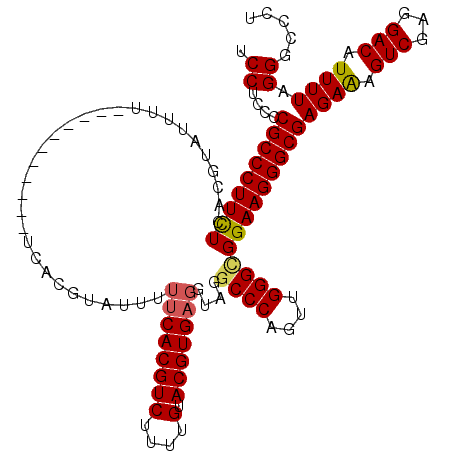
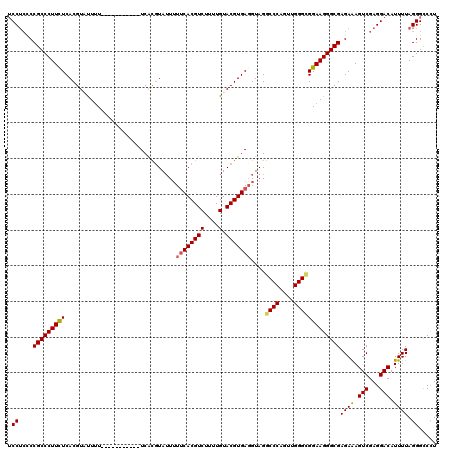
| Location | 3,005,607 – 3,005,715 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -40.59 |
| Consensus MFE | -32.47 |
| Energy contribution | -34.35 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005607 108 - 22407834 AGGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAAAAGACGUGAA-AAUAGGUGA-----------AUAGGACGUGAGAAGGGCGGUGUGGA .((((........))))((.((...(.(((((((((((....(((.(((((..((((((.......)))))).-..)))))..-----------.)))...))).))))))))).)).)) ( -38.50) >DroSec_CAF1 13138 109 - 1 AGGACCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAGAAGACGUGAAAAAUACGUGA-----------AAAAGACGUGAAAAGGGCGGCGAGGA (((((........)))))...((((.((((((((..((((....)))).....((((((.......(((((.....)))))..-----------.....)))))).)))))))).)))). ( -38.74) >DroEre_CAF1 13068 109 - 1 AAGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCUCCCAACUGGGCCUACGUCACGUACAAAAGACGUGGAAAAUACACGA-----------AAAAUACGUGAGAAGGGCGGGUAGGA .....((((....(((...)))...((((((((((..(((....)))......(((((((.......((((.......)))).-----------....))))))))))))))))))))). ( -37.80) >DroYak_CAF1 12771 120 - 1 AGGGCCCUAAAAUGUCCUCGACUCUCUCGCCCUUCCACCCAACUGGGCCUACCUCACGUACACGAGACGUGAAAACUACAUGAAAAACUCGUGAAAAAUACGUGAGAAGGGCGGGGAGGU (((((........)))))...(((((.((((((((..(((....)))......(((((((((((((.((((.......)))).....)))))).....)))))))))))))))))))).. ( -47.30) >consensus AGGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAAAAGACGUGAAAAAUACAUGA___________AAAAGACGUGAGAAGGGCGGGGAGGA (((((........)))))...((((((((((((((.((((....)))).....((((((.......)))))).................................)))))))))))))). (-32.47 = -34.35 + 1.88)

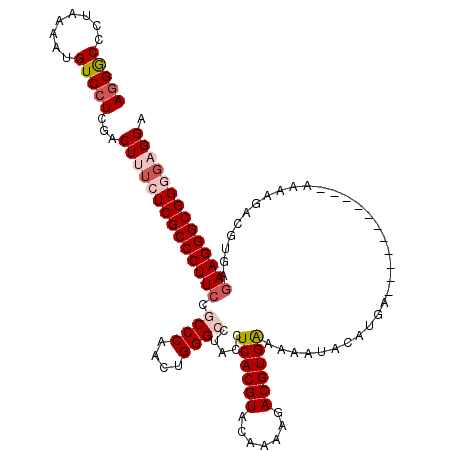
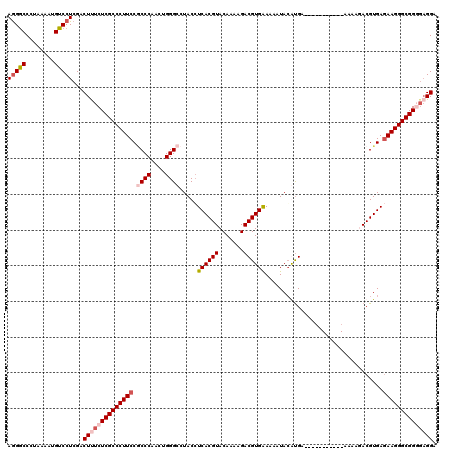
| Location | 3,005,636 – 3,005,752 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -27.20 |
| Energy contribution | -26.82 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3005636 116 - 22407834 UGCCAUGGGAUUACGAAACCCGAGAUGA---UAAUUUAUGAGGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAAAAGACGUGAA-AAUAGG ..(((((((.....(((...((((((((---....)))(((((((........)))))))....)))))...)))..))))..))).((((..((((((.......)))))).-..)))) ( -31.20) >DroSec_CAF1 13167 117 - 1 UGCCAUGGGAUUACGAAACCCGAGAUGA---UAAUUUAUGAGGACCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAGAAGACGUGAAAAAUACG .(((.((((.....(((...((((((((---....)))(((((((........)))))))....)))))...)))..))))....))).....((((((.......))))))........ ( -30.50) >DroEre_CAF1 13097 117 - 1 CGCCAUGGGAUUACGAAACCCGAGAUAA---UAAUUUAUGAAGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCUCCCAACUGGGCCUACGUCACGUACAAAAGACGUGGAAAAUACA .(((.(((((....(((...((((((((---....)))(((.(((........))).)))....)))))...))).)))))....)))(((((((..........)))))))........ ( -26.90) >DroYak_CAF1 12811 120 - 1 UGCCAUGGGAUUACGAAACCCGAGAUAAUAAUAAUUUAUGAGGGCCCUAAAAUGUCCUCGACUCUCUCGCCCUUCCACCCAACUGGGCCUACCUCACGUACACGAGACGUGAAAACUACA .(((.((((.....(((...(((((..((((....))))((((((........)))))).....)))))...)))..))))....))).....((((((.......))))))........ ( -29.50) >consensus UGCCAUGGGAUUACGAAACCCGAGAUAA___UAAUUUAUGAGGGCCCUAAAAUGUCCUCGACUUUCUCGCCCUUCCGCCCAACUGGGCCUACCUCACGUACAAAAGACGUGAAAAAUACA .(((.((((.....(((...((((((((.......)))(((((((........)))))))....)))))...)))..))))....))).....((((((.......))))))........ (-27.20 = -26.82 + -0.38)

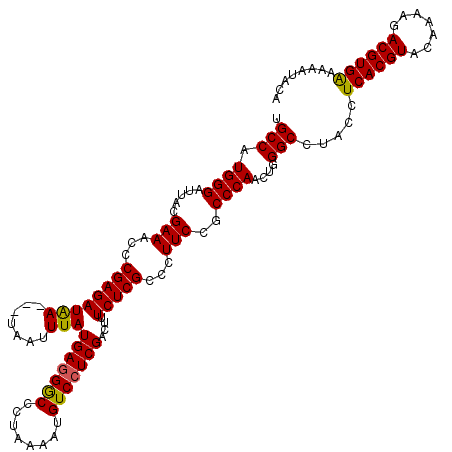
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:41 2006