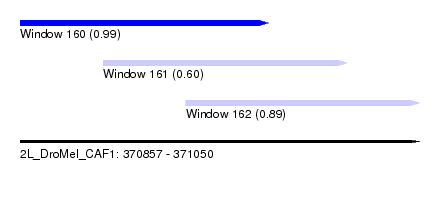
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 370,857 – 371,050 |
| Length | 193 |
| Max. P | 0.992123 |

| Location | 370,857 – 370,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -34.88 |
| Energy contribution | -34.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 370857 120 + 22407834 ACGCAAUGCAAAAAUCUUGUAGCAGGCUGUAACAAGUGGGCGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG ..(((((((.....((((((.((..(((......)))..))...))))))......(....).)))))))..(((((((....))))))).(((((((...........))))))).... ( -35.50) >DroSec_CAF1 38415 120 + 1 ACGCAAUGCAAAAAUCUUGUAGCAGGCUGUAACAAGUGGGCGGGGCAGGGCAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG ..(((((((......(((((.((..(((......)))..))...)))))((....))......)))))))..(((((((....))))))).(((((((...........))))))).... ( -35.70) >DroSim_CAF1 39085 120 + 1 ACGCAAUGCAAAAAUCUUGUAGCAGGCUGUAACAAGUGGGCGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG ..(((((((.....((((((.((..(((......)))..))...))))))......(....).)))))))..(((((((....))))))).(((((((...........))))))).... ( -35.50) >DroEre_CAF1 44953 120 + 1 ACGCAAUGCAAAAAUCUUGUAGCAGGCUGUAACAAGUGGGCGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG ..(((((((.....((((((.((..(((......)))..))...))))))......(....).)))))))..(((((((....))))))).(((((((...........))))))).... ( -35.50) >DroYak_CAF1 39541 120 + 1 ACGCAAUGCAAAAAUCUUGUAGCAGGCUGCAACAAGUGGGCGGGGCAGGACAGGGGCGGGAGAGUAGUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG .((((.((((....((((((..(.(.((((..(........)..))))..).)..))))))......)))).(((((((....))))))).(((((((...........))))))))))) ( -34.10) >consensus ACGCAAUGCAAAAAUCUUGUAGCAGGCUGUAACAAGUGGGCGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCG ..(((((((.....((((((.((..(((......)))..))...))))))......(....).)))))))..(((((((....))))))).(((((((...........))))))).... (-34.88 = -34.92 + 0.04)

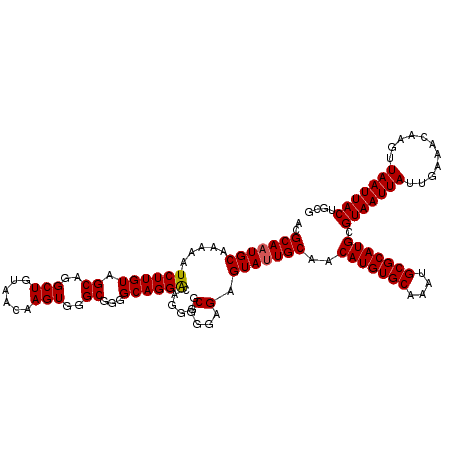
| Location | 370,897 – 371,015 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -35.92 |
| Energy contribution | -36.56 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 370897 118 + 22407834 CGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGAUUCGCU-- ..............((((((......(((((.(((((((....))))))).(((((((...........)))))))))))).(((((((((((....)))))).)).)))..))))))-- ( -37.30) >DroSec_CAF1 38455 118 + 1 CGGGGCAGGGCAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGAU-- ((((((((.((((..((......)).))))..(((((((....))))))).........................))))...(((((((((((....)))))).)).)))..))))..-- ( -39.20) >DroSim_CAF1 39125 118 + 1 CGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGCU-- ..............((((((......(((((.(((((((....))))))).(((((((...........)))))))))))).(((((((((((....)))))).)).)))..))))))-- ( -39.90) >DroEre_CAF1 44993 118 + 1 CGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCGGCGACCGACUCGCU-- ..............((((((......(((((.(((((((....))))))).(((((((...........)))))))))))).(((((((((((....)))))).)).)))..))))))-- ( -39.90) >DroYak_CAF1 39581 120 + 1 CGGGGCAGGACAGGGGCGGGAGAGUAGUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUGCGACGAGUCCAGCGACCGACUCGAUAU (((.((.((((.(..((((....(((.((((.(((((((....))))))).(((((((...........)))))))))))....)))...))))..)..)))).))..)))......... ( -38.50) >consensus CGGGGCAGGACAGGGGCGGGAGAGUAUUGCAACAUGUGCAAAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGCU__ ..............((((((......(((((.(((((((....))))))).(((((((...........)))))))))))).(((((((((((....)))))).)).)))..)))))).. (-35.92 = -36.56 + 0.64)

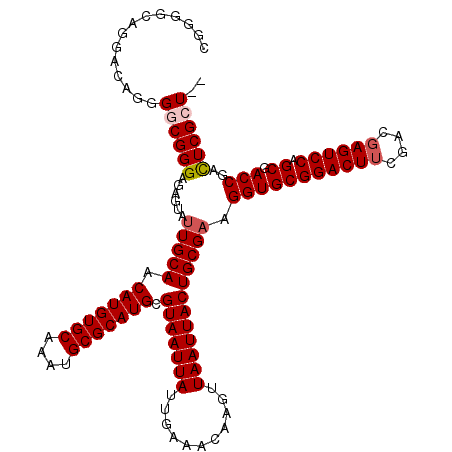
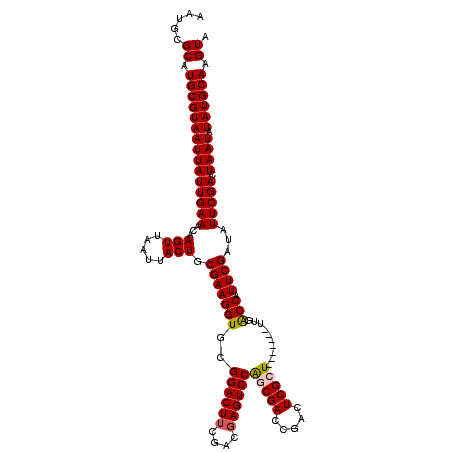
| Location | 370,937 – 371,050 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.99 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 370937 113 + 22407834 AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGAUUCGCU-------UUGACCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....((.(((((((((((((((...(((.....))).(((((((..((((((....))))))(((((.....)))))-------...))).))))...))))).)))).)))))).)). ( -30.60) >DroSec_CAF1 38495 113 + 1 AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGAU-------UUGACCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....(((((((((((((...........))))))).))...(((((((((((....)))))).)).)))...((((.-------((((....))))...)))).......))))..... ( -27.70) >DroSim_CAF1 39165 113 + 1 AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGCU-------UUGACCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....((.(((((((((((((((...(((.....))).(((((((..((((((....))))))(((((.....)))))-------...))).))))...))))).)))).)))))).)). ( -30.60) >DroEre_CAF1 45033 112 + 1 AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCGGCGACCGACUCGCU--------UUGCCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....((.(((((((((((((((...(((.....))).(((((((.(((((((....)))))))((((.....)))).--------..))).))))...))))).)))).)))))).)). ( -31.80) >DroYak_CAF1 39621 120 + 1 AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUGCGACGAGUCCAGCGACCGACUCGAUAUUCGCAUUGACCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....((.(((((((((((((((...(((.....))).(((((((.(((..(((((((((((........))))))....))))))))))).))))...))))).)))).)))))).)). ( -31.70) >consensus AAUGCGCAUGCGUAAUUAUUGAAACAAGUUAAUUACUGCGAAGGUGCGGACUUCGACGAGUCCAGCGACCGACUCGCU_______UUGACCAUUCGAUAUUCGACUAAUAUAUGCAAGUA .....((.(((((((((((((((...(((.....))).(((((((..((((((....))))))(((((.....)))))..........))).))))...))))).)))).)))))).)). (-27.90 = -28.18 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:04 2006