
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,003,979 – 3,004,081 |
| Length | 102 |
| Max. P | 0.787605 |

| Location | 3,003,979 – 3,004,081 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3003979 102 + 22407834 CUUUCUGGCUAGCUUUCUGCUUUCUACUGCCGGCCUGCCGGUCUUCAGCCGCGGCUCUUUCCAACUCCAAGCUCCAAA------------CUGCAGCUCCAACUUCU------ACUCCAA ......(((((((.....))).......(((((....)))))....))))((((......))........((......------------..)).))..........------....... ( -20.50) >DroSec_CAF1 11566 94 + 1 CUUUCUGGCUAGCUUUCUGCUUUCUACUGC--------CGGUCUUCAGCCGAGGCUCUUACCAGCUCCAAGCUUCAAA------------CUGCAGCUCCAACUUCU------ACUCCAA .....(((..(((.....)))..)))((((--------.(((.....)))((((((.............))))))...------------..))))...........------....... ( -17.12) >DroSim_CAF1 10912 102 + 1 CUUUCUGGCUAGCUUUCUGCUUUCUACUGCCGGCCUGCCGGUCUUCAGCCGCGGCUCUUUCCAACUCCAAGCUUCAAA------------CUGCAGCUCCAACUUCU------ACUCCAA ......(((((((.....))).......(((((....)))))....))))((((......))........((......------------..)).))..........------....... ( -20.50) >DroEre_CAF1 11577 114 + 1 CUUUCUGGCUAGCUUUCUGCUUUCUACUGCCGGCCUGCCGGUCUUCAGCUGCGGCUCUUUCCAACUUUAUGCUCCGAACACCAGCUUCAGCUGCAGCUCCAACUUCA------ACUCCAG ....((((..(((.....))).......(((((....)))))....((((((((((..............(((.........)))...)))))))))).........------...)))) ( -28.03) >DroYak_CAF1 11197 120 + 1 CUUUCUGGCUAGCUUUCUGCUUUCUACUGCCGGCCUGCCGGUCUUCAGCUGCGGCUCUUUCCAACUCCGAGCUGCAAACACCAACUUCAGCUGCAGCUCCAACUUCAGCUCAAACUCCAA ......(((((((.....))).......(((((....)))))....))))(((((((...........))))))).............(((((.((......)).))))).......... ( -31.30) >consensus CUUUCUGGCUAGCUUUCUGCUUUCUACUGCCGGCCUGCCGGUCUUCAGCCGCGGCUCUUUCCAACUCCAAGCUCCAAA____________CUGCAGCUCCAACUUCU______ACUCCAA ......(((((((.....))).......(((((....)))))....))))..((......))........(((.((...............)).)))....................... (-14.65 = -14.70 + 0.05)

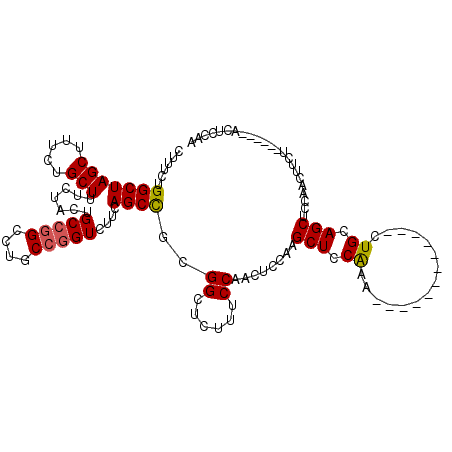
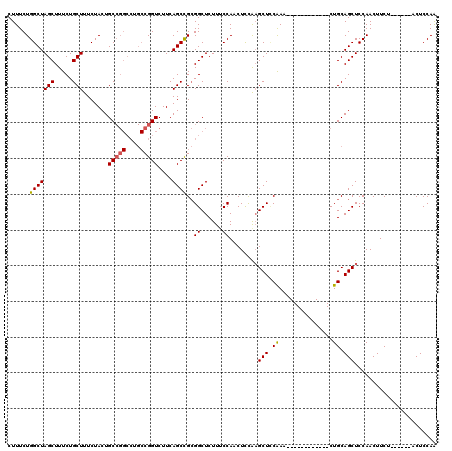
| Location | 3,003,979 – 3,004,081 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -21.53 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3003979 102 - 22407834 UUGGAGU------AGAAGUUGGAGCUGCAG------------UUUGGAGCUUGGAGUUGGAAAGAGCCGCGGCUGAAGACCGGCAGGCCGGCAGUAGAAAGCAGAAAGCUAGCCAGAAAG ((((...------....(((((..((((.(------------(((..(((((((..((....))..))).))))..))))..)))).))))).......(((.....)))..)))).... ( -30.10) >DroSec_CAF1 11566 94 - 1 UUGGAGU------AGAAGUUGGAGCUGCAG------------UUUGAAGCUUGGAGCUGGUAAGAGCCUCGGCUGAAGACCG--------GCAGUAGAAAGCAGAAAGCUAGCCAGAAAG .......------.....((((.(((((.(------------(((..((((.((.(((......))))).))))..))))..--------)))))....(((.....)))..)))).... ( -25.80) >DroSim_CAF1 10912 102 - 1 UUGGAGU------AGAAGUUGGAGCUGCAG------------UUUGAAGCUUGGAGUUGGAAAGAGCCGCGGCUGAAGACCGGCAGGCCGGCAGUAGAAAGCAGAAAGCUAGCCAGAAAG ((((...------....(((((..((((.(------------(((..(((((((..((....))..))).))))..))))..)))).))))).......(((.....)))..)))).... ( -30.50) >DroEre_CAF1 11577 114 - 1 CUGGAGU------UGAAGUUGGAGCUGCAGCUGAAGCUGGUGUUCGGAGCAUAAAGUUGGAAAGAGCCGCAGCUGAAGACCGGCAGGCCGGCAGUAGAAAGCAGAAAGCUAGCCAGAAAG (((((((------(...(((...(((((.(((...((((((.((((((((.....)))((......))....))))).)))))).)))..)))))....)))....))))..)))).... ( -36.80) >DroYak_CAF1 11197 120 - 1 UUGGAGUUUGAGCUGAAGUUGGAGCUGCAGCUGAAGUUGGUGUUUGCAGCUCGGAGUUGGAAAGAGCCGCAGCUGAAGACCGGCAGGCCGGCAGUAGAAAGCAGAAAGCUAGCCAGAAAG (((((((((...(((...((...(((((.(((...((((((.(...((((((((..((....))..))).))))).).)))))).)))..))))).))...))).)))))..)))).... ( -42.00) >consensus UUGGAGU______AGAAGUUGGAGCUGCAG____________UUUGAAGCUUGGAGUUGGAAAGAGCCGCGGCUGAAGACCGGCAGGCCGGCAGUAGAAAGCAGAAAGCUAGCCAGAAAG .................(((((..((((...................(((((((..((....))..))).))))...(.((((....)))))........))))....)))))....... (-21.53 = -22.38 + 0.85)

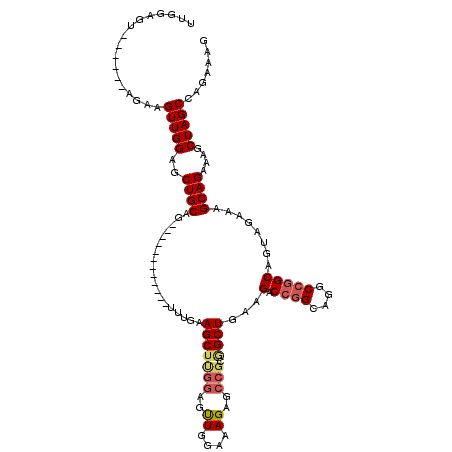
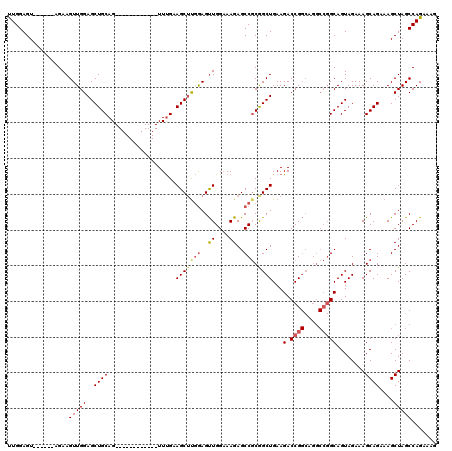
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:35 2006