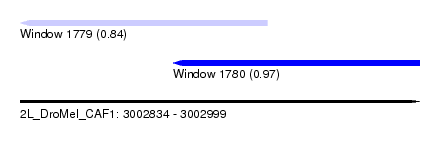
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,002,834 – 3,002,999 |
| Length | 165 |
| Max. P | 0.965899 |

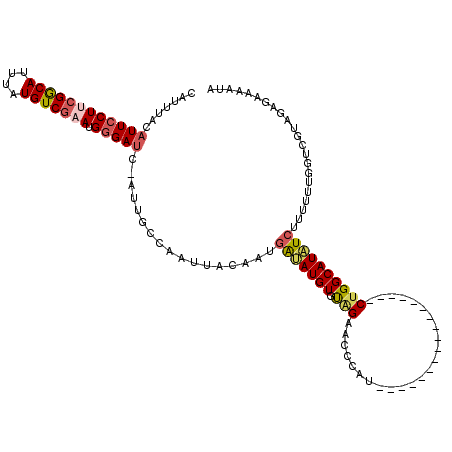
| Location | 3,002,834 – 3,002,936 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3002834 102 - 22407834 CAUUUACAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC-AUUGCCAAUUACAAUGAUAUGUGUAGAACCCAU-----------------CUGGCAUAUCUUUUUUGGUCGUAGAGGAAAUA ..(((((((((((((((((....))))))).))))).-...(((((.......(((((((.((((.....)-----------------))))))))))....))))).)))))....... ( -27.90) >DroSec_CAF1 10376 102 - 1 CAUUUACAUUGCUUCGGCACUUAUGUCGAAUGGGAUC-AUUGCCAAUUAGAAUGAUAUGUGUGGAACCCAU-----------------CUGGCAUAUCAUUUUCGGUCGUAGAGUAAAUA .((((((..(((....))).(((((((((.(((.(..-..).)))....(((((((((((((((...))))-----------------...))))))))))))))).))))).)))))). ( -30.30) >DroSim_CAF1 9735 102 - 1 CAUUUAAAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC-AUUGCCAAUUACAAUGAUAUGUGUAGAACCCAU-----------------CUGGCAUAUCAUUUUCGAUCGUAGAGUAAAUA .(((((.((((((((((((....))))))).))))).-.((((.......((((((((((.((((.....)-----------------))))))))))))).......))))..))))). ( -25.14) >DroEre_CAF1 10385 120 - 1 CAUUUACAUUCCUGAGACAUUUAUGUCGAAUGGGACCGAUUGCCUAUUACAAUUAGAUGUGCAGAUCCCAUUUGGUGUGCAAUUAAAUCUGGCAUCUCUUGCUUGUGCUUAGAGAAAAUA ........(((((((((((..(((..(((((((((....((((.((((.......)))).)))).)))))))))..)))...........((((.....))))))).))))).))).... ( -30.30) >DroYak_CAF1 9954 119 - 1 CAUUUACAUUCCUACGACAUUUAUGUCGAAUGGGAUC-AUUGCCUAUUACAAUGGUAUGUGCAGAUCCCAUUUGGUGUGCAAUUAAAUCGGGCAUCUCUUUCUUGUGCUUAAAGAAAAUG ..............(((((....)))))(((((((((-..((((...(((....))).).)))))))))))).(((((.(.........).)))))..((((((.......))))))... ( -28.90) >consensus CAUUUACAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC_AUUGCCAAUUACAAUGAUAUGUGUAGAACCCAU_________________CUGGCAUAUCUUUUUUGGUCGUAGAGAAAAUA .......((((((((((((....))))))).))))).................(((((((.(((........................))))))))))...................... (-12.64 = -13.24 + 0.60)

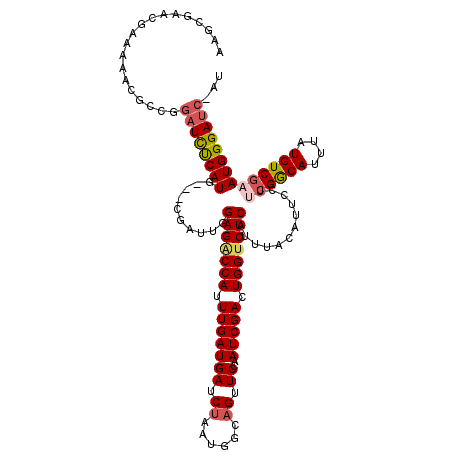
| Location | 3,002,897 – 3,002,999 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.24 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3002897 102 - 22407834 AAGC--------------GGAUUCCAUG---CGAUUGGAGGCCAUUUGAUGAUCUCAUGGCAGUUCCAAUCGACUGGUCUCAUUUACAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC-AU ....--------------.((..(((..---((((((((.(((((..(.....)..)))))...))))))))..)))..))......((((((((((((....))))))).))))).-.. ( -36.70) >DroSec_CAF1 10439 119 - 1 AAGCGAACGAAAAGCGCCGGAUCUCAUGGAUCGAUUGGAGACCAAUUGAUGAUCUAAUGGCAGUUCCAAUCGACUGGUCUCAUUUACAUUGCUUCGGCACUUAUGUCGAAUGGGAUC-AU .(((((..(((....((((((((......(((((((((...))))))))))))))...)))..)))...))).))(((((((((((((((((....)))...)))).))))))))))-.. ( -37.80) >DroSim_CAF1 9798 116 - 1 AAGCGAACGAAAAGCGCCGGAUCUCAUG---CGAUUGGAGACCAUUUGAUGAUCUAAUGGCAGUUCCAAUCGACUGGUCUCAUUUAAAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC-AU ..(((.........))).(((((.....---(((((((((.(((((.(.....).)))))...)))))))))...))))).......((((((((((((....))))))).))))).-.. ( -35.10) >DroEre_CAF1 10465 117 - 1 AAACGAGCGGAAAAUGGCGAAUCCCAUG---CGAUUCGAGACCAUUUGAUGAUCUAAUGUCCGUUCCAAUCGACUGGGCUCAUUUACAUUCCUGAGACAUUUAUGUCGAAUGGGACCGAU ....((.((..((((((((((((.....---.))))))...))))))..)).))....(((.((((((.(((((...(((((..........)))).)......))))).)))))).))) ( -32.60) >DroYak_CAF1 10034 116 - 1 AAACGAGCGAAAGACGCCGGAUCUCAUG---CGAUUCGAGACCAUUUGAUGAUCUAAUGUCAGUUCCAAUCGACUGGUCUCAUUUACAUUCCUACGACAUUUAUGUCGAAUGGGAUC-AU ...((.(((.....)))))((((((((.---......(((((((.(((((((.((......)).))..))))).))))))).............(((((....))))).))))))))-.. ( -36.80) >consensus AAGCGAACGAAAAACGCCGGAUCUCAUG___CGAUUGGAGACCAUUUGAUGAUCUAAUGGCAGUUCCAAUCGACUGGUCUCAUUUACAUUCCUUCGGCAUUUAUGUCGAAUGGGAUC_AU ...................((((((((..........(((((((.(((((((.((......)).))..))))).)))))))............((((((....))))))))))))))... (-26.60 = -27.24 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:30 2006