| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,001,318 – 3,001,438 |
| Length | 120 |
| Max. P | 0.899355 |

| Location | 3,001,318 – 3,001,438 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -37.54 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
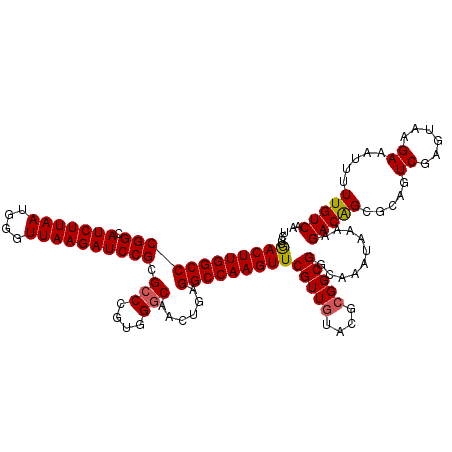
>2L_DroMel_CAF1 3001318 120 + 22407834 UCCUGACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGGCAACUGAGGCCAAGUUCGUUGUACGCGGCGGCAAAUAAAAGACAGCGCAGUCGAGUAAGAAAUUUUUGUCAA ....((((((((((((.(((((((....)))))))))).(((....)))......)))))))))(((((....)))))..........(((((.....((......)).....))))).. ( -39.90) >DroSec_CAF1 8874 120 + 1 UCCUGACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGCCAACUGAGGCCAAGUUCGUUGUACGCGGCGGCAAAUAAAAGACAGCGCAGUCGAGUAAGAAAUUUUUGUCAA ..((.(((((((..((.(((((((....)))))))))((((...(((((......))))).((.(((((....)))))))............)))).))))))).))............. ( -38.60) >DroSim_CAF1 8239 120 + 1 UCCUGACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGGCAACUGAGGCCAAGUUCGUUGUACGCGGCGGCAAAUAAAAGACAGCUCAGUCGAGUAAGAAAUUUUUGUCAA ....((((((((((((.(((((((....)))))))))).(((....)))......))))))))).((((....))))((((((.........((((....))))........)))))).. ( -41.03) >DroEre_CAF1 8868 118 + 1 --CUGACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGGCAACUGAGGCCAAGUUCGUUGUACGCGGCGGAGAAUAAAAGACAGCGCAGUCGAGUAAGAAAUUUUUGUCAA --..((((((((((((.(((((((....)))))))))).(((....)))......)))))))))(((((....)))))..........(((((.....((......)).....))))).. ( -39.90) >DroYak_CAF1 8376 118 + 1 --CCAACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGGCAACUGAGGCCAAGUUCGUUCUACGCGGCGGUGAAUAAAAGACGGCGCAGUCGAGUAAGAAAUUUUUGUCAA --..((((((((((((.(((((((....)))))))))).(((....)))......)))))))))..((((((.((((.(((............))).)))).)).))))........... ( -39.40) >consensus UCCUGACUUGGCCCGGCAUCUUAAUGGGUUAAGAUCCGCGCCCGUGGGCAACUGAGGCCAAGUUCGUUGUACGCGGCGGCAAAUAAAAGACAGCGCAGUCGAGUAAGAAAUUUUUGUCAA ....((((((((((((.(((((((....)))))))))).(((....)))......)))))))))(((((....)))))..........(((((.....((......)).....))))).. (-37.54 = -37.62 + 0.08)

| Location | 3,001,318 – 3,001,438 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -37.34 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
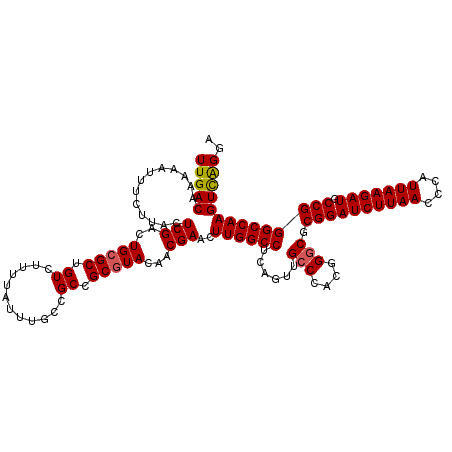
>2L_DroMel_CAF1 3001318 120 - 22407834 UUGACAAAAAUUUCUUACUCGACUGCGCUGUCUUUUAUUUGCCGCCGCGUACAACGAACUUGGCCUCAGUUGCCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUCAGGA (((((.............(((..(((((.((............)).)))))...)))..((((((......(((....))).((((((((((....))))))).)))))))))))))).. ( -36.60) >DroSec_CAF1 8874 120 - 1 UUGACAAAAAUUUCUUACUCGACUGCGCUGUCUUUUAUUUGCCGCCGCGUACAACGAACUUGGCCUCAGUUGGCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUCAGGA (((((.............(((..(((((.((............)).)))))...))).(.(((((......))))).)(((.((((((((((....))))))).))).)))..))))).. ( -38.50) >DroSim_CAF1 8239 120 - 1 UUGACAAAAAUUUCUUACUCGACUGAGCUGUCUUUUAUUUGCCGCCGCGUACAACGAACUUGGCCUCAGUUGCCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUCAGGA (((((..............((((((((.............((((...((.....))....))))))))))))......(((.((((((((((....))))))).))).)))..))))).. ( -35.91) >DroEre_CAF1 8868 118 - 1 UUGACAAAAAUUUCUUACUCGACUGCGCUGUCUUUUAUUCUCCGCCGCGUACAACGAACUUGGCCUCAGUUGCCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUCAG-- .((((.............(((..(((((.((............)).)))))...)))..((((((......(((....))).((((((((((....))))))).))))))))))))).-- ( -36.20) >DroYak_CAF1 8376 118 - 1 UUGACAAAAAUUUCUUACUCGACUGCGCCGUCUUUUAUUCACCGCCGCGUAGAACGAACUUGGCCUCAGUUGCCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUUGG-- ......................((((((.((............)).))))))..(.(((((((((......(((....))).((((((((((....))))))).)))))))))))).)-- ( -39.50) >consensus UUGACAAAAAUUUCUUACUCGACUGCGCUGUCUUUUAUUUGCCGCCGCGUACAACGAACUUGGCCUCAGUUGCCCACGGGCGCGGAUCUUAACCCAUUAAGAUGCCGGGCCAAGUCAGGA (((((.............(((..(((((.((............)).)))))...)))..((((((......(((....))).((((((((((....))))))).)))))))))))))).. (-33.16 = -33.24 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:27 2006