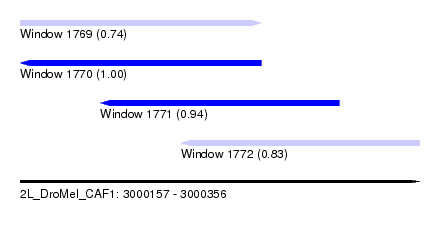
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,000,157 – 3,000,356 |
| Length | 199 |
| Max. P | 0.998578 |

| Location | 3,000,157 – 3,000,277 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.58 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3000157 120 + 22407834 AGGCUCUUCAGCUAUUGAAAAAACAUUGAAUCAAUACUUUUUUACGUGUACCAAGCCCACGUCAGUCGACUAAAAAUCCAUGGGCCUCGGCCCACGGAAAAUUAACUCAUUGCCCCCUCU .(((...((.(((...((((((..((((...))))..))))))(((((.........))))).))).)).......(((.(((((....))))).))).............)))...... ( -25.30) >DroSec_CAF1 7686 120 + 1 AGGCUCCUCAACUAUUGAAAAAACAUUGAAUCAAUACUUUUCUACGUGUACCAAGCCCACGUCAGUCGGCUGAAAAUCCAUGGGCCUCGGCCCACGGAAAAUUAACUCAUUGCCCCCACU .(((........((((((............)))))).........(.((....((((.((....)).)))).....(((.(((((....))))).)))......)).)...)))...... ( -27.80) >DroSim_CAF1 7053 120 + 1 AGGCUCUUCAACUAUUGAAAAAACAUUGAAUCAAUACUUUUCUACGUGUACCAAGCCCACGUCAGUCGGCUGAAAAUCCAUGGGCCUCGGCCCACGGAAAAUUAACUCAUUGCCCCCACU .(((..(((...((((((............)))))).................((((.((....)).)))))))..(((.(((((....))))).))).............)))...... ( -28.20) >DroEre_CAF1 7652 119 + 1 AGACUCUUUGGCUAUUGAA-AAACAUUGAAUCAAUACUUCUCUAUGUGUACCAAGCCCACGUCAGUCGGCUGAAAAUCCAUGGUCCUCGGCCCACGGGAAAUUAACUCAUUGCCCCCACU .(((.....((((..((..-..((((.(((.......)))...))))....))))))...))).((.((((((..((.....))..)))))).))(((.(((......))).)))..... ( -23.70) >DroYak_CAF1 7159 119 + 1 CGACUCUUCAGCGAUUGAA-AAACAUUGAAUCAAUACUUCUCUACGUGUACCAAGCCCACGUCAGUCGGCUGAAAAUCCAUGGGUCUCGGCCCACGGAAAAUUAACUCAUUGCCCCCACU ......((((((((((((.-....((((...)))).........((((.........)))))))))).))))))..(((.(((((....))))).)))...................... ( -28.50) >consensus AGGCUCUUCAGCUAUUGAAAAAACAUUGAAUCAAUACUUUUCUACGUGUACCAAGCCCACGUCAGUCGGCUGAAAAUCCAUGGGCCUCGGCCCACGGAAAAUUAACUCAUUGCCCCCACU .(((..((((((((((((.......(((.....((((........))))..))).......))))).)))))))..(((.(((((....))))).))).............)))...... (-21.38 = -22.58 + 1.20)

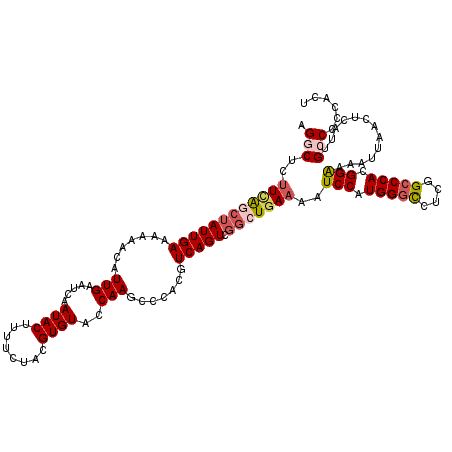
| Location | 3,000,157 – 3,000,277 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -34.86 |
| Energy contribution | -35.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3000157 120 - 22407834 AGAGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUUAGUCGACUGACGUGGGCUUGGUACACGUAAAAAAGUAUUGAUUCAAUGUUUUUUCAAUAGCUGAAGAGCCU .(((((((((.(((((((((..(((((((((....)))))))))..........(((.(((((..(...)..))))).....)))))))))))).)))))))))....(((....))).. ( -42.40) >DroSec_CAF1 7686 120 - 1 AGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGAAAAGUAUUGAUUCAAUGUUUUUUCAAUAGUUGAGGAGCCU .....((((..(((((((((((((((((..(((((.((((((.....((((....)))))))))))))))..))))..))))...))))))))).....((((((.....)))))))))) ( -42.10) >DroSim_CAF1 7053 120 - 1 AGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGAAAAGUAUUGAUUCAAUGUUUUUUCAAUAGUUGAAGAGCCU .....((((..(((((((((((((((((..(((((.((((((.....((((....)))))))))))))))..))))..))))...))))))))).....((((((.....)))))))))) ( -42.50) >DroEre_CAF1 7652 119 - 1 AGUGGGGGCAAUGAGUUAAUUUCCCGUGGGCCGAGGACCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACAUAGAGAAGUAUUGAUUCAAUGUUU-UUCAAUAGCCAAAGAGUCU ..((((((.(((......))).)))(((..(((((..(((((.....((((....))))))))).)))))..)))...(((((((((((...))))))))-))).....)))........ ( -34.70) >DroYak_CAF1 7159 119 - 1 AGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGACCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGAGAAGUAUUGAUUCAAUGUUU-UUCAAUCGCUGAAGAGUCG ((((((((((.(((((((((((((((((..(((((.((((((.....((((....)))))))))))))))..))))....)))).))))))))).)))))-.....)))))......... ( -40.60) >consensus AGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGAAAAGUAUUGAUUCAAUGUUUUUUCAAUAGCUGAAGAGCCU ......(((..(((((((((((((((((..(((((.((((((.....((((....)))))))))))))))..))))..))))...))))))))).....((((((.....))))))))). (-34.86 = -35.66 + 0.80)

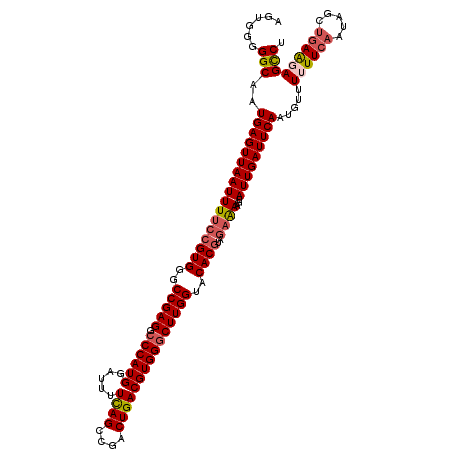
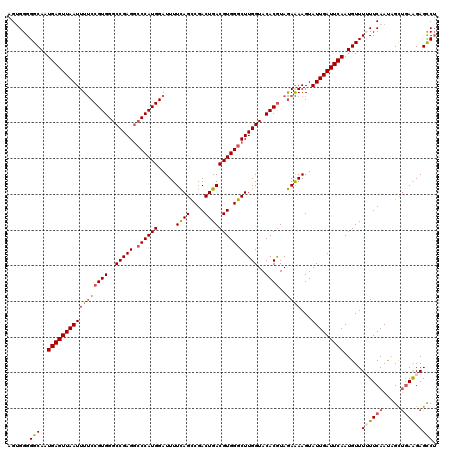
| Location | 3,000,197 – 3,000,316 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3000197 119 - 22407834 CCCGAGGAGAAUAGAGCAAUUUUCUGGAUCGA-AGGGAAAAGAGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUUAGUCGACUGACGUGGGCUUGGUACACGUAAA (((..(((((((......)))))))...((..-..))......))).......(((..(((.(((((((((....))))))))).....)))..))).(((((..(...)..)))))... ( -36.80) >DroSec_CAF1 7726 120 - 1 CCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGAAAGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGA ((((((((...........)))))))).((....)).....(((...((.............(((((((((....))))))))).....((((.((....)).))))..)).)))..... ( -41.60) >DroSim_CAF1 7093 119 - 1 CCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGA ((((((((...........)))))))).....-..((((((.....(((.....)))..))))))(((..(((((.((((((.....((((....)))))))))))))))..)))..... ( -40.50) >DroEre_CAF1 7691 119 - 1 CCCUAAGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUCCCGUGGGCCGAGGACCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACAUAGA (((...((((((......))))))))).....-.((((((......(((.....)))..))))))(((..(((((..(((((.....((((....))))))))).)))))..)))..... ( -34.20) >DroYak_CAF1 7198 119 - 1 CCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGACCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGA ((((((((...........)))))))).....-..((((((.....(((.....)))..))))))(((..(((((.((((((.....((((....)))))))))))))))..)))..... ( -40.50) >consensus CCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA_AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCAUGGAUUUUCAGCCGACUGACGUGGGCUUGGUACACGUAGA ((((((((...........))))))))........((((((.....(((.....)))..))))))(((..(((((.((((((.....((((....)))))))))))))))..)))..... (-35.72 = -36.20 + 0.48)

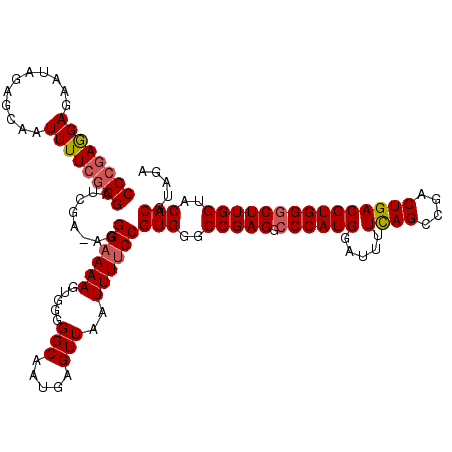
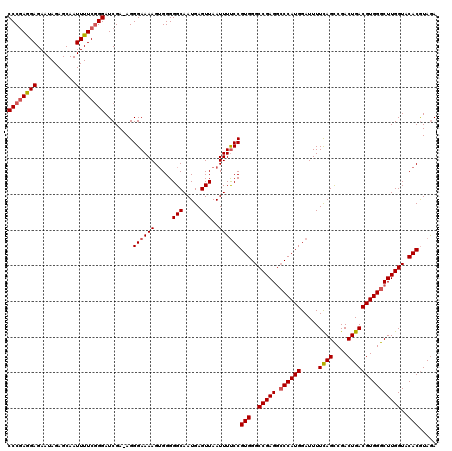
| Location | 3,000,237 – 3,000,356 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -31.40 |
| Energy contribution | -32.64 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3000237 119 - 22407834 AAAUGGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCGAGGAGAAUAGAGCAAUUUUCUGGAUCGA-AGGGAAAAGAGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCA ...(((((..(((((((((..((((.....)))).))))))(((.(((((((((..((.((((((...((..-..))...)))))).....))..)).))))))).))))))...))))) ( -39.00) >DroSec_CAF1 7766 120 - 1 AAAUGGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGAAAGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCA ...(((((..(((..((...(((((.((...)).)))))(((((((((...........))))))))).(....)((((((.....(((.....)))..))))))))..)))...))))) ( -40.50) >DroSim_CAF1 7133 119 - 1 AAAUGGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCA ...(((((..(((..((...(((((.((...)).)))))(((((((((...........)))))))))....-..((((((.....(((.....)))..))))))))..)))...))))) ( -39.90) >DroEre_CAF1 7731 119 - 1 AAAUAGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCUAAGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUCCCGUGGGCCGAGGACCA .....(((....((((..(((((((.((...)).)))((((((...((((((......))))))....((..-..))......)))))).....))))....))))...)))........ ( -31.40) >DroYak_CAF1 7238 119 - 1 AAAUAGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA-AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGACCCA ............(((((...(.(((.((........((.(((((((((...........))))))))).)).-..((((((.....(((.....)))..)))))))).))))..).)))) ( -32.40) >consensus AAAUGGGCAAGGUGGGCGUAGCGCUGACAAAGUGGGCGUUCCCGAGGAGAAUAGAGCAAUUUUCGGGAUCGA_AGGGAAAAGUGGGGGCAAUGAGUUAAUUUUCCGUGGGCCGAGGCCCA ...(((((..(((..((...(((((.((...)).)))))(((((((((...........))))))))).......((((((.....(((.....)))..))))))))..)))...))))) (-31.40 = -32.64 + 1.24)

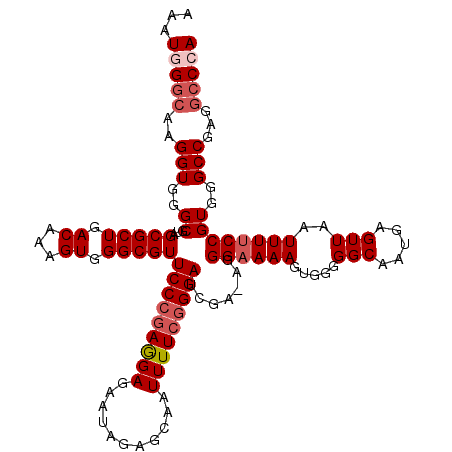
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:22 2006