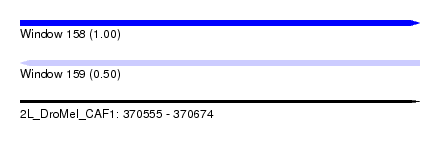
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 370,555 – 370,674 |
| Length | 119 |
| Max. P | 0.998383 |

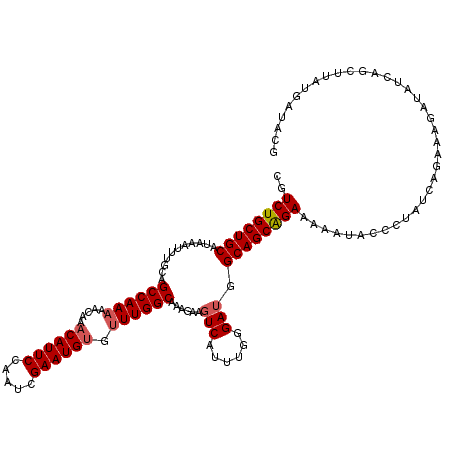
| Location | 370,555 – 370,674 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 370555 119 + 22407834 CGUCUGCUGCAUAAAUUUGCAGCCAAAAACAAACAUUCCAAUCGAAUGUGUUUGGCAAAGAAGUCAUUUGGGAUGGCAGCAGAAAAAUACCCUAUCAGAAAGAUAUCAGCUUAUGAUAC- ..((((((((...........((((((.....((((((.....)))))).))))))......(((......))).)))))))).........(((((..(((.......))).))))).- ( -31.70) >DroSec_CAF1 38107 120 + 1 CGUCUGCUGCAUAAAUUUGCAGCCAAAAACAAACAUUCCAAUCGAAUGUGUUUGGCAAAGGAGUCAUUUGGGAUGGCAGCAGAAAAAUACCCUAUCAGAAAGAUACCAGCUUAUGAUAUG ..((((((((...........((((((.....((((((.....)))))).))))))......(((......))).)))))))).........(((((..(((.......))).))))).. ( -31.80) >DroSim_CAF1 38777 120 + 1 CGUCUGCUGCAUAAAUUUGCAGCCAAAAACAAACAUUCCAAUCGAAUGUGUUUGGCAAAGGAGUCAUUUGGGAUGGCAGCAGAAAAAUACCCUAUCUGAAAGAAAUCAGCUUAUGAUAUG ..((((((((...........((((((.....((((((.....)))))).))))))......(((......))).))))))))............((((......))))........... ( -30.80) >DroEre_CAF1 44642 118 + 1 CGUCUGCUGCAUAAAUUUGCAGCCAA-AACAAACAUUCCAAUCGAAUGCGUUUGGCAAGGAAGUCAUUUGGGAUGGCAGCAGAAGAAUACCCUAUCAAAAAGAUGU-AGCUUAGCACACG .((..(((((((...(((((.(((((-(.....(((((.....)))))..))))))......((((((...)))))).))))).((........))......))))-)))...))..... ( -29.90) >DroYak_CAF1 39236 120 + 1 CGUCUGCUGCAUAAAUUUGCAGCCAAAAACAAACAUUCCAAUCGAAUGUGUUUGGCAAAGAAGUCAUUUGCGAAGGCAGCGGAAAAAUACCCUAACAUCAAGAUAUCAACUAGGGAUACG ..((((((((.....((((((((((((.....((((((.....)))))).))))))...(....)...)))))).))))))))......(((((................)))))..... ( -36.69) >consensus CGUCUGCUGCAUAAAUUUGCAGCCAAAAACAAACAUUCCAAUCGAAUGUGUUUGGCAAAGAAGUCAUUUGGGAUGGCAGCAGAAAAAUACCCUAUCAGAAAGAUAUCAGCUUAUGAUACG ..((((((((...........((((((.....((((((.....)))))).))))))......(((......))).))))))))..................................... (-26.84 = -27.28 + 0.44)

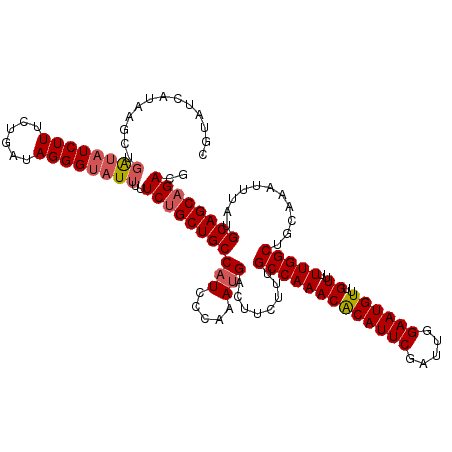
| Location | 370,555 – 370,674 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 370555 119 - 22407834 -GUAUCAUAAGCUGAUAUCUUUCUGAUAGGGUAUUUUUCUGCUGCCAUCCCAAAUGACUUCUUUGCCAAACACAUUCGAUUGGAAUGUUUGUUUUUGGCUGCAAAUUUAUGCAGCAGACG -............((((((((......))))))))..(((((((((((.....)))........((((((.((((((.....)))))).....))))))...........)))))))).. ( -28.80) >DroSec_CAF1 38107 120 - 1 CAUAUCAUAAGCUGGUAUCUUUCUGAUAGGGUAUUUUUCUGCUGCCAUCCCAAAUGACUCCUUUGCCAAACACAUUCGAUUGGAAUGUUUGUUUUUGGCUGCAAAUUUAUGCAGCAGACG .............((((((((......))))))))..(((((((((((.....)))........((((((.((((((.....)))))).....))))))...........)))))))).. ( -28.30) >DroSim_CAF1 38777 120 - 1 CAUAUCAUAAGCUGAUUUCUUUCAGAUAGGGUAUUUUUCUGCUGCCAUCCCAAAUGACUCCUUUGCCAAACACAUUCGAUUGGAAUGUUUGUUUUUGGCUGCAAAUUUAUGCAGCAGACG .(((((.....((((......))))....)))))...(((((((((((.....)))........((((((.((((((.....)))))).....))))))...........)))))))).. ( -31.10) >DroEre_CAF1 44642 118 - 1 CGUGUGCUAAGCU-ACAUCUUUUUGAUAGGGUAUUCUUCUGCUGCCAUCCCAAAUGACUUCCUUGCCAAACGCAUUCGAUUGGAAUGUUUGUU-UUGGCUGCAAAUUUAUGCAGCAGACG ..........(((-..(((.....)))..))).....(((((((((((.....)))........((((((.((((((.....))))))....)-)))))...........)))))))).. ( -28.60) >DroYak_CAF1 39236 120 - 1 CGUAUCCCUAGUUGAUAUCUUGAUGUUAGGGUAUUUUUCCGCUGCCUUCGCAAAUGACUUCUUUGCCAAACACAUUCGAUUGGAAUGUUUGUUUUUGGCUGCAAAUUUAUGCAGCAGACG .(((.(((((((..(....)..))..)))))))).....((((((...................((((((.((((((.....)))))).....))))))((((......)))))))).)) ( -26.50) >consensus CGUAUCAUAAGCUGAUAUCUUUCUGAUAGGGUAUUUUUCUGCUGCCAUCCCAAAUGACUUCUUUGCCAAACACAUUCGAUUGGAAUGUUUGUUUUUGGCUGCAAAUUUAUGCAGCAGACG .............((((((((......))))))))..(((((((((((.....)))........(((((((((((((.....))))))..))..)))))...........)))))))).. (-24.74 = -25.42 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:01 2006