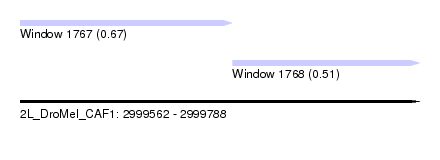
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,999,562 – 2,999,788 |
| Length | 226 |
| Max. P | 0.670991 |

| Location | 2,999,562 – 2,999,682 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -29.45 |
| Energy contribution | -29.53 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2999562 120 + 22407834 AACCGAAAAUAGUUCGCGAAACGCAGCAGGCGACGCGUCUUUUUGGCUCUCGAAAAGGGCAUGAAAACUCGCGAACUGUGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ........(((((((((((..(((.....)))..((.((((((((.....))))))))))........)))))))))))............................(((....)))... ( -31.80) >DroSec_CAF1 7042 120 + 1 AACCGAAAAUAGUUCGCGAAACGCAGCAGGCGACGCGUCUUUUGGGCUCUCGAAAAGGGCAUGAAAACUCGCGAACUGGGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ............((((((((.(((.((.(....)))((((((((((((((.....((..(.(((....))).)..))))))))..))))))))..............))).)).)))))) ( -30.70) >DroSim_CAF1 6417 120 + 1 AACCGAAAAUAGUUCGCGAAACGCAGCAGGCGACGCGUCUUUUGGGCUCUCGAAAAGGGCAUGAAAACUCGCGAACUGCGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ............((((((((.(((.((((....((((..((((.((((((.....))))).).))))..))))..))))..((.................)).....))).)).)))))) ( -31.83) >DroEre_CAF1 7024 120 + 1 AACCGAAAAUAGUUCGCGAAACGCAGCAGGCGACGCGUCUUUUGGGCUCUCGAAAAGGGCAUGAAAACUCGCGAUCUGCGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ............((((((((.(((.(((((...((((..((((.((((((.....))))).).))))..)))).)))))..((.................)).....))).)).)))))) ( -31.83) >DroYak_CAF1 6502 120 + 1 AAACGAAAAUAGUUCGCGAAACGCGGCAGGCGACGCGUCUUUUGGGCUCUCAAAAAGGGCAUGAAAACUCGCGAACUGCGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ............((((((((.((((((..(((.((((..((((.((((((.....))))).).))))..))))...))).)))...........(.....)......))).)).)))))) ( -32.20) >consensus AACCGAAAAUAGUUCGCGAAACGCAGCAGGCGACGCGUCUUUUGGGCUCUCGAAAAGGGCAUGAAAACUCGCGAACUGCGGCCAAUAAAAGAUACAUACAGGAAAAAGCGAUUGCGCGAA ............((((((((.(((.((((....((((..((((.((((((.....))))).).))))..))))..))))..((.................)).....))).)).)))))) (-29.45 = -29.53 + 0.08)

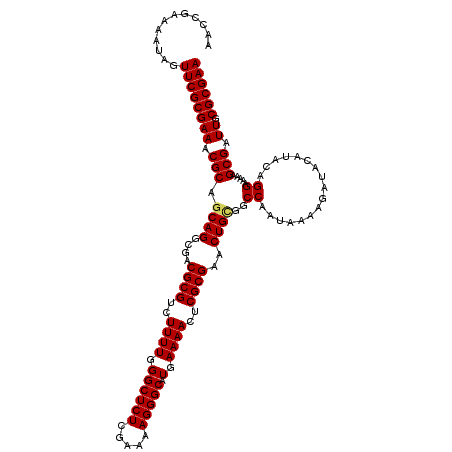
| Location | 2,999,682 – 2,999,788 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -21.90 |
| Energy contribution | -21.34 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2999682 106 + 22407834 UUCUCGGCUGCGUGUGCGCUGCAAAUUUGCUCGUGCGGAGC----------AAAAACAAACGAAAAUAGCCGGAUGAAC----AAAUACAUCCAGUGCGAAGAAUAAUCUACGCUGUAUA .....(((((((....)))......(((((((.....))))----------))).............))))(((((...----.....)))))...(((.(((....))).)))...... ( -30.10) >DroSec_CAF1 7162 110 + 1 UUCUCGGCUGCGUGUGCGCUGCAAAUUUGCUCGUGCGGAGC----------AAAAACAAACGAAAAUAGCCGGAUGAACAAACAAAUACAUCCAGUGCGAAGAAUAAUCUGCGCUGUAUA .....(((((((....)))......(((((((.....))))----------))).............))))(((((............)))))(((((..(((....))))))))..... ( -29.70) >DroSim_CAF1 6537 110 + 1 UUCUCGGCUGCGUGAGCGCUGCAAAUUUGCUCGUGGGGAGC----------AAAAACAAACGAAAAUAGCCGGAUGAACAAACAAAUACAUCCAGUGCGAAGAAUAAUCUGCGCUGUAUA .....((((((....))........(((((((.....))))----------))).............))))(((((............)))))(((((..(((....))))))))..... ( -30.80) >DroEre_CAF1 7144 109 + 1 UUCUCGGCUGCGUGUGCGCUGCAAAUUUGCUCGUGCGUAGC----------AAAAACAAACGAAAAUAGCCG-AUGAACAAACAAAUACAGGCAGUGUGAAGAAUAAUCUGCGCUGUUUA ((((((((((((....)))......(((((((....).)))----------))).............)))))-).)))...........(((((((((..(((....)))))))))))). ( -29.70) >DroYak_CAF1 6622 119 + 1 UUCUCGGCUGCGUGUGCGCUGCAAAUUUGCUCGUGCGGAGCAAAGACAAGCAAAAACAAACGAAAAUAGCCG-AUGAACAAACAAAUACGGCUAGUGUGAAGAAUAAUCUGCGCUGUUUA ((((((((((((....)))(((...(((((((.....))))))).....)))...............)))))-).))).........(((((((((((.....)))..))).)))))... ( -30.60) >consensus UUCUCGGCUGCGUGUGCGCUGCAAAUUUGCUCGUGCGGAGC__________AAAAACAAACGAAAAUAGCCGGAUGAACAAACAAAUACAUCCAGUGCGAAGAAUAAUCUGCGCUGUAUA (((.((((((((....))).........((((.....))))..........................)))))...)))...........((.((((((..(((....))))))))).)). (-21.90 = -21.34 + -0.56)

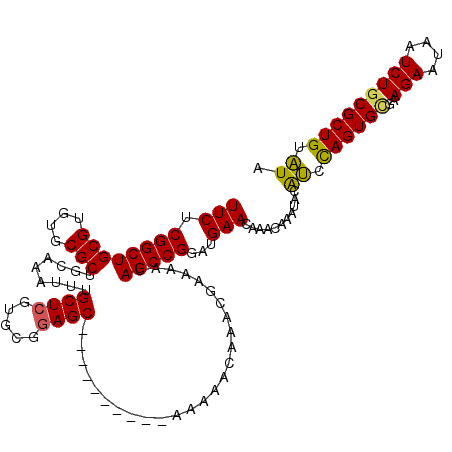
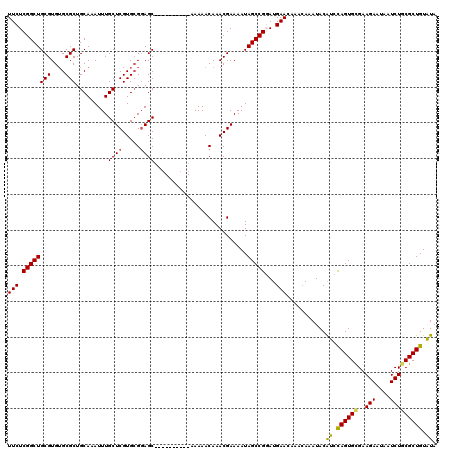
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:18 2006