
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,993,672 – 2,993,792 |
| Length | 120 |
| Max. P | 0.550218 |

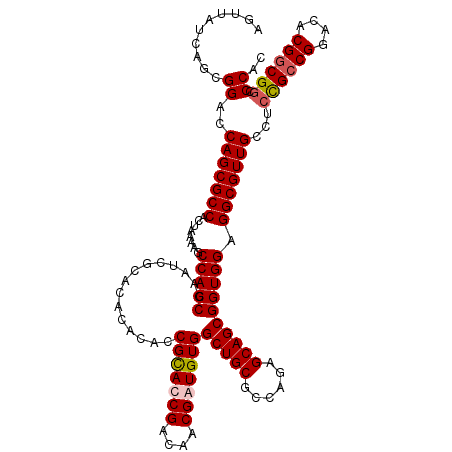
| Location | 2,993,672 – 2,993,792 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -44.58 |
| Energy contribution | -45.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
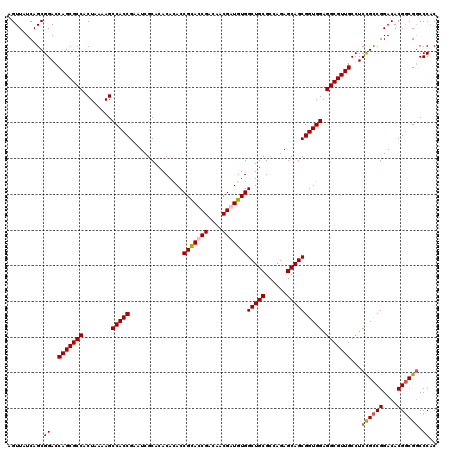
>2L_DroMel_CAF1 2993672 120 + 22407834 AGUUAUCAGCGGACCAGCGCCACUAAAAGCCACCGAAUCGCACACACACCGCACCGACAACGAUGUGGCUGCGCCAGAGCAGCGGUGGAGGCGUUGCCUCCGUCGGACACGUCGGCCCAC .(((....(((((.(((((((........(((((...............((((.((....)).))))(((((......)))))))))).)))))))..)))))..)))............ ( -41.60) >DroSec_CAF1 5427 120 + 1 AGUUAUCAGCGGACCAGCGCCACUAAAAGCCACCGAAUCGCACACACACCGCAUCGACAACGAUGUGGCUGCGCCAGAGCAGCGGUGGAGGCGUUGCCUCCGCCGGACACGGCGUCCCAC ........(.(((((((((((........(((((...............(((((((....)))))))(((((......)))))))))).))))))).....((((....))))))))).. ( -51.20) >DroSim_CAF1 5409 120 + 1 AGUUAUCAGCGGACCAGCGCCACUAAAAGCCACCGAAUCGCACACACACCGUAUCGACAACGAUGUGGCUGCGCCAGAGCAGCGGUGGAGGCGUUGCCUCCGCCGGACACGGCGGCCCAC ..........((..(((((((........(((((..........((((.(((.......))).))))(((((......)))))))))).)))))))...((((((....)))))).)).. ( -49.00) >DroYak_CAF1 5443 120 + 1 AGUUAUCAGCGGACCAGCGCCACUAAGAGCCACCGAAUCGCACACAUACCGCACCGAAAACGAUGUGGCUGCGCCAGAGCAGCGGUGGAGGCGUUGCCUCUGCCGGACACGGCGGCCCCC ..........((..(((((((........(((((...............((((.((....)).))))(((((......)))))))))).))))))))).((((((....))))))..... ( -44.10) >consensus AGUUAUCAGCGGACCAGCGCCACUAAAAGCCACCGAAUCGCACACACACCGCACCGACAACGAUGUGGCUGCGCCAGAGCAGCGGUGGAGGCGUUGCCUCCGCCGGACACGGCGGCCCAC ..........((..(((((((........(((((...............(((((((....)))))))(((((......)))))))))).)))))))...((((((....)))))).)).. (-44.58 = -45.20 + 0.62)

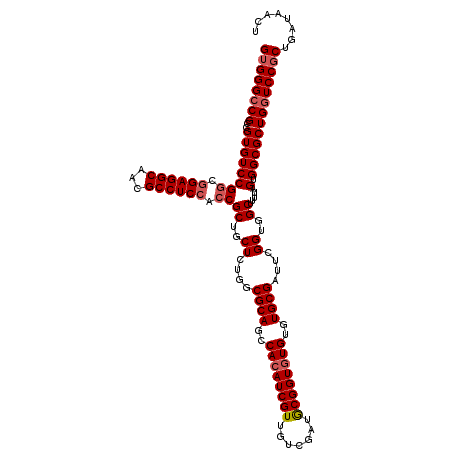
| Location | 2,993,672 – 2,993,792 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -55.60 |
| Consensus MFE | -51.66 |
| Energy contribution | -52.72 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
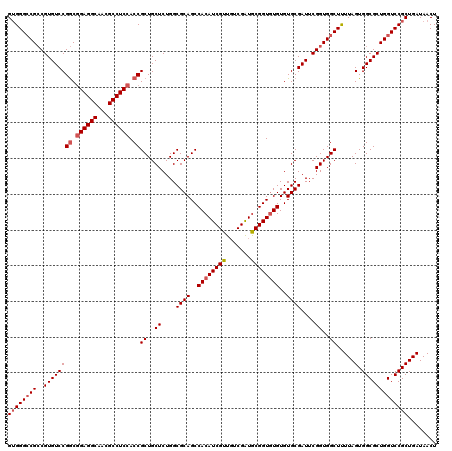
>2L_DroMel_CAF1 2993672 120 - 22407834 GUGGGCCGACGUGUCCGACGGAGGCAACGCCUCCACCGCUGCUCUGGCGCAGCCACAUCGUUGUCGGUGCGGUGUGUGUGCGAUUCGGUGGCUUUUAGUGGCGCUGGUCCGCUGAUAACU ((((((((..((((((...((((((...))))))...((..((....((((..((((((((.......))))))))..))))....))..)).....).)))))))))))))........ ( -55.00) >DroSec_CAF1 5427 120 - 1 GUGGGACGCCGUGUCCGGCGGAGGCAACGCCUCCACCGCUGCUCUGGCGCAGCCACAUCGUUGUCGAUGCGGUGUGUGUGCGAUUCGGUGGCUUUUAGUGGCGCUGGUCCGCUGAUAACU ((((...(((((((((((.((((((...)))))).))((..((....((((..((((((((.......))))))))..))))....))..)).....).))))).)))))))........ ( -54.00) >DroSim_CAF1 5409 120 - 1 GUGGGCCGCCGUGUCCGGCGGAGGCAACGCCUCCACCGCUGCUCUGGCGCAGCCACAUCGUUGUCGAUACGGUGUGUGUGCGAUUCGGUGGCUUUUAGUGGCGCUGGUCCGCUGAUAACU ((((((((..((((((((.((((((...)))))).))((..((....((((..((((((((.......))))))))..))))....))..)).....).)))))))))))))........ ( -58.40) >DroYak_CAF1 5443 120 - 1 GGGGGCCGCCGUGUCCGGCAGAGGCAACGCCUCCACCGCUGCUCUGGCGCAGCCACAUCGUUUUCGGUGCGGUAUGUGUGCGAUUCGGUGGCUCUUAGUGGCGCUGGUCCGCUGAUAACU ..(((((((((.((((((..(((((...)))))..)))........((((((((.(((((....))))).)))...)))))))).)))))))))((((((((....).)))))))..... ( -55.00) >consensus GUGGGCCGCCGUGUCCGGCGGAGGCAACGCCUCCACCGCUGCUCUGGCGCAGCCACAUCGUUGUCGAUGCGGUGUGUGUGCGAUUCGGUGGCUUUUAGUGGCGCUGGUCCGCUGAUAACU ((((((((..((((((((.((((((...)))))).))((..((....((((..((((((((.......))))))))..))))....))..)).....).)))))))))))))........ (-51.66 = -52.72 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:13 2006